7NL0
 
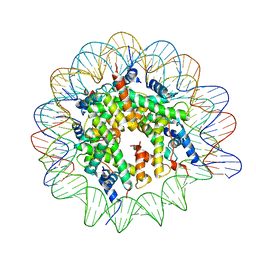 | | Cryo-EM structure of the Lin28B nucleosome core particle | | Descriptor: | DNA (131-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Roberts, G.A, Ozkan, B, Gachulincova, I, O Dwyer, M.R, Hall-Ponsele, E, Saxena, M, Robinson, P.J, Soufi, A. | | Deposit date: | 2021-02-19 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Dissecting OCT4 defines the role of nucleosome binding in pluripotency.
Nat.Cell Biol., 23, 2021
|
|
8OEO
 
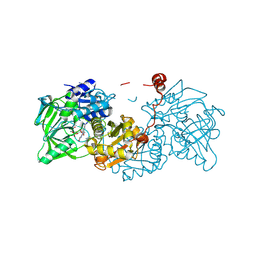 | | Aspergillus niger ferulic acid decarboxylase (Fdc) V186C-A296C (DB4) variant in complex with prenylated flavin | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Roberts, G.W, Leys, D. | | Deposit date: | 2023-03-10 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Aspergillus niger ferulic acid decarboxylase (Fdc) V186C-A296C (DB4) variant in complex with prenylated flavin
To Be Published
|
|
8OED
 
 | |
8OEH
 
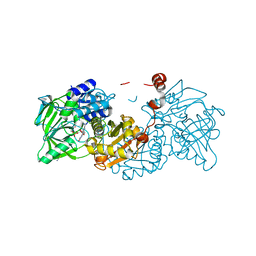 | | Aspergillus niger ferulic acid decarboxylase (Fdc) C122-S261C (DB3) variant in complex with prenylated flavin | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Roberts, G.W, Leys, D. | | Deposit date: | 2023-03-10 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Aspergillus niger ferulic acid decarboxylase (Fdc) C122-S261C (DB3) variant in complex with prenylated flavin
To Be Published
|
|
8CRD
 
 | |
2WCJ
 
 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) with (10E,12Z)-tetradecadien-1-ol | | Descriptor: | (10E,12Z)-tetradeca-10,12-dien-1-ol, GENERAL ODORANT-BINDING PROTEIN 1, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
2WMA
 
 | |
2WMB
 
 | | Structural and thermodynamic consequences of cyclization of peptide ligands for the recruitment site of cyclin A | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, LINEAR RKLFD, ... | | Authors: | Robertson, G.F, Endicott, J.A, Noble, M.E.M, McDonnell, J.M. | | Deposit date: | 2009-06-30 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Thermodynamic Consequences of Cyclization of Peptide Ligands for the Recruitment Site of Cyclin A
To be Published
|
|
2WCH
 
 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) with bombykal | | Descriptor: | (10E,12Z)-hexadeca-10,12-dienal, GENERAL ODORANT-BINDING PROTEIN 1, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
2WCM
 
 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) with (10E)-hexadecen-12-yn-1-ol | | Descriptor: | (10E)-hexadec-10-en-12-yn-1-ol, GENERAL ODORANT-BINDING PROTEIN 1, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
2WCK
 
 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) without ligand | | Descriptor: | GENERAL ODORANT-BINDING PROTEIN 1, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
2WCL
 
 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) with (8E, 10Z)-hexadecadien-1-ol | | Descriptor: | (8E,10Z)-HEXADECA-8,10-DIEN-1-OL, GENERAL ODORANT-BINDING PROTEIN 1, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
2WC6
 
 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) with bombykol and water to Arg 110 | | Descriptor: | GENERAL ODORANT-BINDING PROTEIN 1, HEXADECA-10,12-DIEN-1-OL, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-09 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
2WC5
 
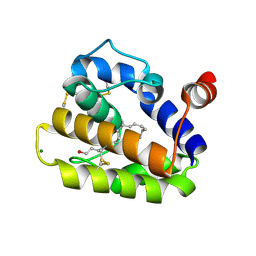 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) | | Descriptor: | GENERAL ODORANT-BINDING PROTEIN 1, HEXADECA-10,12-DIEN-1-OL, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-09 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
3MSP
 
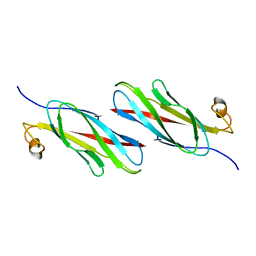 | | MOTILE MAJOR SPERM PROTEIN (MSP) OF ASCARIS SUUM, NMR, 20 STRUCTURES | | Descriptor: | MAJOR SPERM PROTEIN | | Authors: | Haaf, A, Leclaire III, L, Roberts, G, Kent, H.M, Roberts, T.M, Stewart, M, Neuhaus, D. | | Deposit date: | 1998-09-10 | | Release date: | 1999-04-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the motile major sperm protein (MSP) of Ascaris suum - evidence for two manganese binding sites and the possible role of divalent cations in filament formation.
J.Mol.Biol., 284, 1998
|
|
3IVF
 
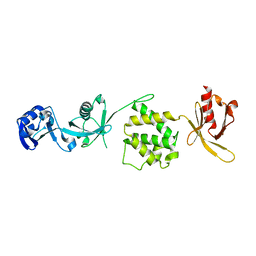 | | Crystal structure of the talin head FERM domain | | Descriptor: | Talin-1 | | Authors: | Elliott, P.R, Goult, B.T, Bate, N, Grossmann, J.G, Roberts, G.C.K, Critchley, D.R, Barsukov, I.L. | | Deposit date: | 2009-09-01 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The Structure of the talin head reveals a novel extended conformation of the FERM domain
Structure, 18, 2010
|
|
1B1C
 
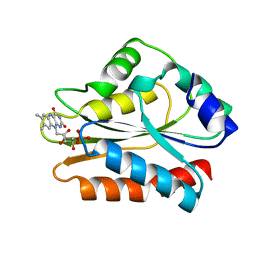 | | CRYSTAL STRUCTURE OF THE FMN-BINDING DOMAIN OF HUMAN CYTOCHROME P450 REDUCTASE AT 1.93A RESOLUTION | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, PROTEIN (NADPH-CYTOCHROME P450 REDUCTASE) | | Authors: | Zhao, Q, Modi, S, Smith, G, Paine, M, Mcdonagh, P.D, Wolf, C.R, Tew, D, Lian, L.-Y, Roberts, G.C.K, Driessen, H.P.C. | | Deposit date: | 1998-11-19 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of the FMN-binding domain of human cytochrome P450 reductase at 1.93 A resolution.
Protein Sci., 8, 1999
|
|
1RHO
 
 | | STRUCTURE OF RHO GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR | | Descriptor: | RHO GDP-DISSOCIATION INHIBITOR 1, SULFATE ION | | Authors: | Keep, N.H, Moody, P.C.E, Roberts, G.C.K. | | Deposit date: | 1996-10-12 | | Release date: | 1997-10-15 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A modulator of rho family G proteins, rhoGDI, binds these G proteins via an immunoglobulin-like domain and a flexible N-terminal arm.
Structure, 5, 1997
|
|
2IGG
 
 | | DETERMINATION OF THE SOLUTION STRUCTURES OF DOMAINS II AND III OF PROTEIN G FROM STREPTOCOCCUS BY 1H NMR | | Descriptor: | PROTEIN G | | Authors: | Lian, L.-Y, Derrick, J.P, Sutcliffe, M.J, Yang, J.C, Roberts, G.C.K. | | Deposit date: | 1992-08-26 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Determination of the solution structures of domains II and III of protein G from Streptococcus by 1H nuclear magnetic resonance.
J.Mol.Biol., 228, 1992
|
|
2M5D
 
 | |
2M5C
 
 | |
1CFP
 
 | |
1AO8
 
 | | DIHYDROFOLATE REDUCTASE COMPLEXED WITH METHOTREXATE, NMR, 21 STRUCTURES | | Descriptor: | DIHYDROFOLATE REDUCTASE, METHOTREXATE | | Authors: | Gargaro, A.R, Soteriou, A, Frenkiel, T.A, Bauer, C.J, Birdsall, B, Polshakov, V.I, Barsukov, I.L, Roberts, G.C.K, Feeney, J. | | Deposit date: | 1997-07-22 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the complex of Lactobacillus casei dihydrofolate reductase with methotrexate.
J.Mol.Biol., 277, 1998
|
|
2IGH
 
 | | DETERMINATION OF THE SOLUTION STRUCTURES OF DOMAINS II AND III OF PROTEIN G FROM STREPTOCOCCUS BY 1H NMR | | Descriptor: | PROTEIN G | | Authors: | Lian, L.-Y, Derrick, J.P, Sutcliffe, M.J, Yang, J.C, Roberts, G.C.K. | | Deposit date: | 1992-08-26 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Determination of the solution structures of domains II and III of protein G from Streptococcus by 1H nuclear magnetic resonance.
J.Mol.Biol., 228, 1992
|
|
1ZW3
 
 | | Vinculin Head (0-258) in Complex with the Talin Rod residues 1630-1652 | | Descriptor: | Talin 1, Vinculin | | Authors: | Gingras, A.R, Ziegler, W.H, Barsukov, I.L, Roberts, G.C, Critchley, D.R, Emsley, J. | | Deposit date: | 2005-06-03 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mapping and consensus sequence identification for multiple vinculin binding sites within the talin rod
J.Biol.Chem., 280, 2005
|
|
