6TGW
 
 | |
1AFE
 
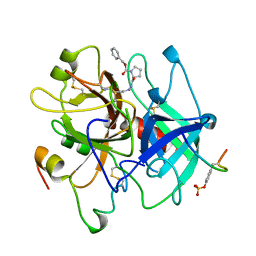 | | HUMAN ALPHA-THROMBIN INHIBITION BY CBZ-PRO-AZALYS-ONP | | Descriptor: | 2-[N'-(4-AMINO-BUTYL)-HYDRAZINOCARBONYL]-PYRROLIDINE-1-CARBOXYLIC ACID BENZYL ESTER, 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ... | | Authors: | De Simone, G, Balliano, G, Milla, P, Gallina, C, Giordano, C, Tarricone, C, Rizzi, M, Bolognesi, M, Ascenzi, P. | | Deposit date: | 1997-03-06 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human alpha-thrombin inhibition by the highly selective compounds N-ethoxycarbonyl-D-Phe-Pro-alpha-azaLys p-nitrophenyl ester and N-carbobenzoxy-Pro-alpha-azaLys p-nitrophenyl ester: a kinetic, thermodynamic and X-ray crystallographic study.
J.Mol.Biol., 269, 1997
|
|
1AE8
 
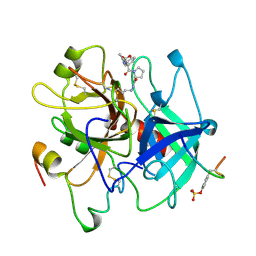 | | HUMAN ALPHA-THROMBIN INHIBITION BY EOC-D-PHE-PRO-AZALYS-ONP | | Descriptor: | 1-ETHOXYCARBONYL-D-PHE-PRO-2(4-AMINOBUTYL)HYDRAZINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ... | | Authors: | De Simone, G, Balliano, G, Milla, P, Gallina, C, Giordano, C, Tarricone, C, Rizzi, M, Bolognesi, M, Ascenzi, P. | | Deposit date: | 1997-03-06 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human alpha-thrombin inhibition by the highly selective compounds N-ethoxycarbonyl-D-Phe-Pro-alpha-azaLys p-nitrophenyl ester and N-carbobenzoxy-Pro-alpha-azaLys p-nitrophenyl ester: a kinetic, thermodynamic and X-ray crystallographic study.
J.Mol.Biol., 269, 1997
|
|
3ZUK
 
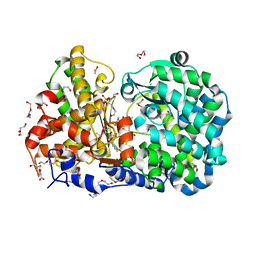 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS ZINC METALLOPROTEASE ZMP1 IN COMPLEX WITH INHIBITOR | | Descriptor: | 2,2',2''-NITRILOTRIETHANOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Ferraris, D.M, Sbardella, D, Petrera, A, Marini, S, Amstutz, B, Coletta, M, Sander, P, Rizzi, M. | | Deposit date: | 2011-07-19 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Mycobacterium Tuberculosis Zinc-Dependent Metalloprotease-1 (Zmp1), a Metalloprotease Involved in Pathogenicity.
J.Biol.Chem., 286, 2011
|
|
6R76
 
 | |
6R77
 
 | |
6T3E
 
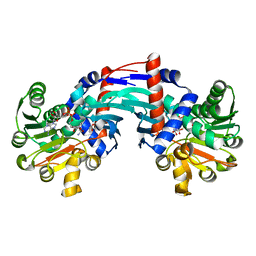 | | Structure of Thermococcus litoralis Delta(1)-pyrroline-2-carboxylate reductase in complex with NADH and L-proline | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DELTA1-pyrroline-2-carboxylate reductase, PROLINE | | Authors: | Ferraris, D.M, Miggiano, R, Ferrario, E, Rizzi, M. | | Deposit date: | 2019-10-10 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Thermococcus litoralis Delta1-pyrroline-2-carboxylate reductase in complex with NADH and L-proline.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5HKI
 
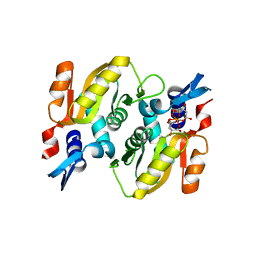 | | Crystal structure of Mycobacterium tuberculosis H37Rv orotate phosphoribosyltransferase in complex with Fe(III) dicitrate | | Descriptor: | Iron(III) dicitrate, Orotate phosphoribosyltransferase | | Authors: | Donini, S, Ferraris, D.M, Bolognesi, G, Rizzi, M. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural investigations on orotate phosphoribosyltransferase from Mycobacterium tuberculosis, a key enzyme of the de novo pyrimidine biosynthesis.
Sci Rep, 7, 2017
|
|
3ZQJ
 
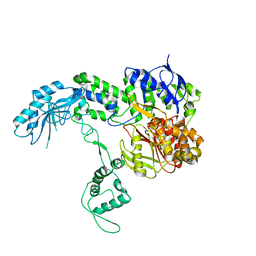 | | Mycobacterium tuberculosis UvrA | | Descriptor: | UVRABC SYSTEM PROTEIN A, ZINC ION | | Authors: | Rossi, F, Khanduja, J.S, Bortoluzzi, A, Houghton, J, Sander, P, Guthlein, C, Davis, E.O, Springer, B, Bottger, E.C, Relini, A, Penco, A, Muniyappa, K, Rizzi, M. | | Deposit date: | 2011-06-09 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Biological and Structural Characterization of Mycobacterium Tuberculosis Uvra Provides Novel Insights Into its Mechanism of Action
Nucleic Acids Res., 39, 2011
|
|
5HKF
 
 | | Crystal structure of Mycobacterium tuberculosis H37Rv orotate phosphoribosyltransferase in complex with 5-phospho-alpha-D-ribosyl 1-diphosphate (PRPP) | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Orotate phosphoribosyltransferase | | Authors: | Donini, S, Ferraris, D.M, Bolognesi, G, Rizzi, M. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural investigations on orotate phosphoribosyltransferase from Mycobacterium tuberculosis, a key enzyme of the de novo pyrimidine biosynthesis.
Sci Rep, 7, 2017
|
|
2HUF
 
 | | Crystal structure of Aedes aegypti alanine glyoxylate aminotransferase | | Descriptor: | 1-BUTANOL, Alanine glyoxylate aminotransferase | | Authors: | Han, Q, Robinson, H, Gao, Y.G, Vogelaar, N, Wilson, S.R, Rizzi, M, Li, J. | | Deposit date: | 2006-07-26 | | Release date: | 2006-09-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Aedes aegypti Alanine Glyoxylate Aminotransferase.
J.Biol.Chem., 281, 2006
|
|
2HUI
 
 | | Crystal structure of Aedes aegypti alanine glyoxylate aminotransferase in complex with glyoxylic acid | | Descriptor: | Alanine glyoxylate aminotransferase, GLYOXYLIC ACID | | Authors: | Han, Q, Robinson, H, Gao, Y.G, Vogelaar, N, Wilson, S.R, Rizzi, M, Li, J. | | Deposit date: | 2006-07-26 | | Release date: | 2006-09-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Aedes aegypti Alanine Glyoxylate Aminotransferase.
J.Biol.Chem., 281, 2006
|
|
2HUU
 
 | | Crystal structure of Aedes aegypti alanine glyoxylate aminotransferase in complex with alanine | | Descriptor: | 1-BUTANOL, ALANINE, Alanine glyoxylate aminotransferase | | Authors: | Han, Q, Robinson, H, Gao, Y.G, Vogelaar, N, Wilson, S.R, Rizzi, M, Li, J. | | Deposit date: | 2006-07-27 | | Release date: | 2006-09-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Aedes aegypti Alanine Glyoxylate Aminotransferase.
J.Biol.Chem., 281, 2006
|
|
5FHZ
 
 | |
5HKL
 
 | |
1EBC
 
 | | SPERM WHALE MET-MYOGLOBIN:CYANIDE COMPLEX | | Descriptor: | CYANIDE ION, PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Rosano, C, Ascenzi, P, Rizzi, M, Losso, R, Bolognesi, M. | | Deposit date: | 1999-03-04 | | Release date: | 1999-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyanide binding to Lucina pectinata hemoglobin I and to sperm whale myoglobin: an x-ray crystallographic study.
Biophys.J., 77, 1999
|
|
1DY9
 
 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (inhibitor I) | | Descriptor: | N-(tert-butoxycarbonyl)-L-alpha-glutamyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70), ... | | Authors: | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
1DY8
 
 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (inhibitor II) | | Descriptor: | N-[(benzyloxy)carbonyl]-L-isoleucyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70) | | Authors: | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
4ZYE
 
 | |
4ZYD
 
 | |
4ZYH
 
 | |
4ZYG
 
 | |
1DXP
 
 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (apo structure) | | Descriptor: | GLYCEROL, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70), ... | | Authors: | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | Deposit date: | 2000-01-13 | | Release date: | 2001-01-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
2XSR
 
 | | Crystal structure of wild type Acinetobacter radioresistens catechol 1,2 dioxygenase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, CATECHOL 1,2 DIOXYGENASE, FE (III) ION | | Authors: | Micalella, C, Martignon, S, Bruno, S, Rizzi, M. | | Deposit date: | 2010-09-30 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Crystallography, Mass Spectrometry and Single Crystal Microspectrophotometry: A Multidisciplinary Characterization of Catechol 1,2 Dioxygenase.
Biochim.Biophys.Acta, 1814, 2011
|
|
3ZTV
 
 | | Structure of Haemophilus influenzae NAD nucleotidase (NadN) | | Descriptor: | ADENOSINE, GLYCEROL, NAD NUCLEOTIDASE, ... | | Authors: | Garavaglia, S, Bruzzone, S, Cassani, C, Canella, L, Allegrone, G, Sturla, L, Mannino, E, Millo, E, De Flora, A, Rizzi, M. | | Deposit date: | 2011-07-12 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The High-Resolution Crystal Structure of Periplasmic Haemophilus Influenzae Nad Nucleotidase Reveals a Novel Enzymatic Function of Human Cd73 Related to Nad Metabolism.
Biochem.J., 441, 2012
|
|
