3UVR
 
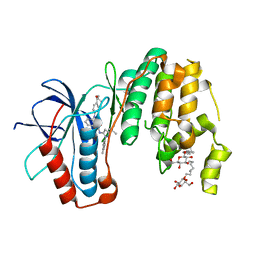 | | Human p38 MAP Kinase in Complex with KM064 | | Descriptor: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-{3-[(5-oxo-6,7,8,9-tetrahydro-5H-benzo[7]annulen-2-yl)amino]phenyl}urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Richters, A, Mayer-Wrangowski, S.C, Gruetter, C, Rauh, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: |
|
|
8FDA
 
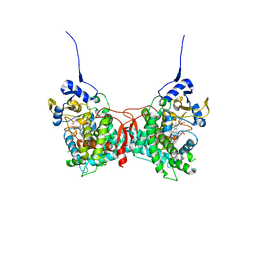 | | Human Cytochrome P450 17A1 in complex with steroidal isonitrile inhibitor | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Steroid 17-alpha-hydroxylase/17,20 lyase, [(1~{R})-1-[(3~{S},5~{S},8~{R},9~{S},10~{S},13~{S},17~{R})-3-methanoyloxy-10,13-dimethyl-1,2,3,4,5,6,7,8,9,11,12,13,14,15,16,17-hexadecahydrocyclopenta[a]phenanthren-17-yl]ethyl]-methylidyne-azanium | | Authors: | Richard, A.M, Scott, E.E. | | Deposit date: | 2022-12-02 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selective steroidogenic cytochrome P450 haem iron ligation by steroid-derived isonitriles.
Commun Chem, 6, 2023
|
|
4LFH
 
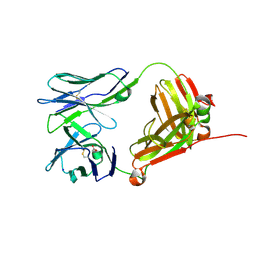 | | Crystal Structure of 9C2 TCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9C2 TCR delta chain, 9C2 TCR gamma chain, ... | | Authors: | Uldrich, A.P, Le Nours, J, Pellicci, D.G, Gras, S, Rossjohn, J, Godfrey, D.I. | | Deposit date: | 2013-06-26 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CD1d-lipid antigen recognition by the gamma delta TCR.
Nat.Immunol., 14, 2013
|
|
4L8M
 
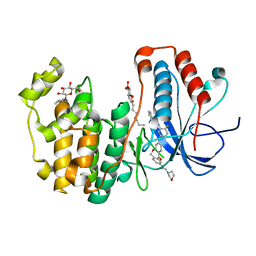 | | Human p38 MAP kinase in complex with a Dibenzoxepinone | | Descriptor: | Mitogen-activated protein kinase 14, N-[2-fluoro-5-({9-[2-(morpholin-4-yl)ethoxy]-11-oxo-6,11-dihydrodibenzo[b,e]oxepin-3-yl}amino)phenyl]benzamide, octyl beta-D-glucopyranoside | | Authors: | Richters, A, Mayer-Wrangowski, S.C, Gruetter, C, Rauh, D. | | Deposit date: | 2013-06-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Metabolically Stable Dibenzo[b,e]oxepin-11(6H)-ones as Highly Selective p38 MAP Kinase Inhibitors: Optimizing Anti-Cytokine Activity in Human Whole Blood.
J.Med.Chem., 56, 2013
|
|
4LHU
 
 | | Crystal Structure of 9C2 TCR bound to CD1d | | Descriptor: | (15Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]tetracos-15-enamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Uldrich, A.P, Le Nours, J, Pellicci, D.G, Gras, S, Rossjohn, J, Godfrey, D.I. | | Deposit date: | 2013-07-01 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | CD1d-lipid antigen recognition by the gamma delta TCR.
Nat.Immunol., 14, 2013
|
|
4DX0
 
 | | Structure of the 14-3-3/PMA2 complex stabilized by a pyrazole derivative | | Descriptor: | 14-3-3-like protein E, 4-[(4R)-4-(4-nitrophenyl)-6-oxidanylidene-3-phenyl-1,4-dihydropyrrolo[3,4-c]pyrazol-5-yl]benzoic acid, N.plumbaginifolia H+-translocating ATPase mRNA | | Authors: | Richter, A, Rose, R, Hedberg, C, Waldmann, H, Ottmann, C. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An Optimised Small-Molecule Stabiliser of the 14-3-3-PMA2 Protein-Protein Interaction.
Chemistry, 18, 2012
|
|
4FIC
 
 | |
4O2P
 
 | | Kinase domain of cSrc in complex with a substituted pyrazolopyrimidine | | Descriptor: | 1-[(2R)-2-chloro-2-phenylethyl]-6-{[2-(morpholin-4-yl)ethyl]sulfanyl}-N-phenyl-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Richters, A, Rauh, D. | | Deposit date: | 2013-12-17 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Combining X-ray Crystallography and Molecular Modeling toward the Optimization of Pyrazolo[3,4-d]pyrimidines as Potent c-Src Inhibitors Active in Vivo against Neuroblastoma.
J.Med.Chem., 58, 2015
|
|
4R3E
 
 | |
2N6Z
 
 | | Solution structure of the salicylate-loaded ArCP from yersiniabactin synthetase | | Descriptor: | HMWP2 nonribosomal peptide synthetase, S-{2-[(N-{(2R)-2-hydroxy-3,3-dimethyl-4-[(trihydroxy-lambda~5~-phosphanyl)oxy]butanoyl}-beta-alanyl)amino]ethyl} 2-hydroxybenzene-1-carbothioate | | Authors: | Goodrich, A.C, Harden, B.J, Frueh, D.P. | | Deposit date: | 2015-08-31 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Nonribosomal Peptide Synthetase Carrier Protein Loaded with Its Substrate Reveals Transient, Well-Defined Contacts.
J.Am.Chem.Soc., 137, 2015
|
|
2N6Y
 
 | |
3E2U
 
 | | Crystal structure of the zink-knuckle 2 domain of human CLIP-170 in complex with CAP-Gly domain of human dynactin-1 (p150-GLUED) | | Descriptor: | CAP-Gly domain-containing linker protein 1, Dynactin subunit 1, ZINC ION | | Authors: | Weisbrich, A, Honnappa, S, Capitani, G, Steinmetz, M.O. | | Deposit date: | 2008-08-06 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-function relationship of CAP-Gly domains
Nat.Struct.Mol.Biol., 14, 2007
|
|
4RVT
 
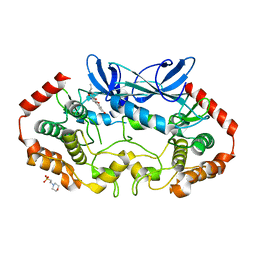 | | MAP4K4 in complex with a pyridin-2(1H)-one derivative | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-hexanoyl-4-hydroxy-5-(4-hydroxyphenyl)pyridin-2(1H)-one, Mitogen-activated protein kinase kinase kinase kinase 4 | | Authors: | Richters, A, Becker, C, Kleine, S, Rauh, D. | | Deposit date: | 2014-11-27 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Neuritogenic Militarinone-Inspired 4-Hydroxypyridones Target the Stress Pathway Kinase MAP4K4.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4R3F
 
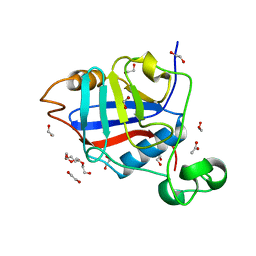 | | Structure of the spliceosomal peptidyl-prolyl cis-trans isomerase Cwc27 from Chaetomium thermophilum | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETATE ION, ... | | Authors: | Ulrich, A, Wahl, M.C. | | Deposit date: | 2014-08-15 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and evolution of the spliceosomal peptidyl-prolyl cis-trans isomerase Cwc27.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6SJY
 
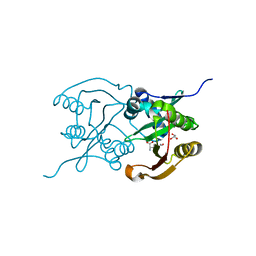 | | Diaminobutyrate acetyltransferase EctA from Paenibacillus lautus in complex with its product ADABA | | Descriptor: | (2~{S})-4-acetamido-2-azanyl-butanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Richter, A.A, Kobus, S, Czech, L, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2019-08-14 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The architecture of the diaminobutyrate acetyltransferase active site provides mechanistic insight into the biosynthesis of the chemical chaperone ectoine.
J.Biol.Chem., 295, 2020
|
|
6SLK
 
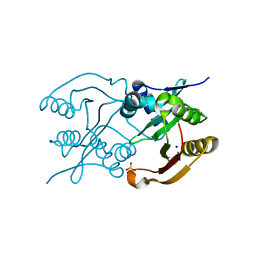 | | Diaminobutyrate acetyltransferase EctA from Paenibacillus lautus | | Descriptor: | L-2,4-diaminobutyric acid acetyltransferase, SODIUM ION, SULFATE ION | | Authors: | Richter, A.A, Kobus, S, Czech, L, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2019-08-20 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The architecture of the diaminobutyrate acetyltransferase active site provides mechanistic insight into the biosynthesis of the chemical chaperone ectoine.
J.Biol.Chem., 295, 2020
|
|
5F5S
 
 | |
6SL8
 
 | | Diaminobutyrate acetyltransferase EctA from Paenibacillus lautus in complex with its substrate L-2,4-diaminobutyric acid (DAB) | | Descriptor: | 2,4-DIAMINOBUTYRIC ACID, GLYCEROL, L-2,4-diaminobutyric acid acetyltransferase, ... | | Authors: | Richter, A.A, Kobus, S, Czech, L, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2019-08-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The architecture of the diaminobutyrate acetyltransferase active site provides mechanistic insight into the biosynthesis of the chemical chaperone ectoine.
J.Biol.Chem., 295, 2020
|
|
6SK1
 
 | | Diaminobutyrate acetyltransferase EctA from Paenibacillus lautus in complex with coenzyme A | | Descriptor: | ACETATE ION, COENZYME A, L-2,4-diaminobutyric acid acetyltransferase | | Authors: | Richter, A.A, Kobus, S, Czech, L, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2019-08-14 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The architecture of the diaminobutyrate acetyltransferase active site provides mechanistic insight into the biosynthesis of the chemical chaperone ectoine.
J.Biol.Chem., 295, 2020
|
|
6SLL
 
 | | Diaminobutyrate acetyltransferase EctA from Paenibacillus lautus in complex with its substrate L-2,4-diaminobutyric acid (DAB) and coenzyme A | | Descriptor: | 2,4-DIAMINOBUTYRIC ACID, COENZYME A, L-2,4-diaminobutyric acid acetyltransferase, ... | | Authors: | Richter, A.A, Kobus, S, Czech, L, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2019-08-20 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The architecture of the diaminobutyrate acetyltransferase active site provides mechanistic insight into the biosynthesis of the chemical chaperone ectoine.
J.Biol.Chem., 295, 2020
|
|
5EN6
 
 | |
5EN7
 
 | |
5EN8
 
 | |
5F5U
 
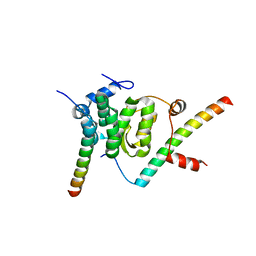 | | Crystal structure of the Snu23-Prp38-MFAP1(217-258) complex of Chaetomium thermophilum | | Descriptor: | Prp38, Putative uncharacterized protein, Zinc finger domain-containing protein | | Authors: | Ulrich, A.K.C, Seeger, M, Bartlick, N, Wahl, M.C. | | Deposit date: | 2015-12-04 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.748 Å) | | Cite: | Scaffolding in the Spliceosome via Single alpha Helices.
Structure, 24, 2016
|
|
5F5V
 
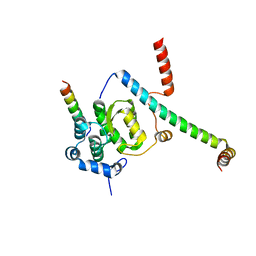 | | Crystal structure of the Snu23-Prp38-MFAP1(217-296) complex of Chaetomium thermophilum | | Descriptor: | Prp38, Putative uncharacterized protein, Zinc finger domain-containing protein | | Authors: | Ulrich, A.K.C, Seeger, M, Bartlick, N, Wahl, M.C. | | Deposit date: | 2015-12-04 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Scaffolding in the Spliceosome via Single alpha Helices.
Structure, 24, 2016
|
|
