2N4O
 
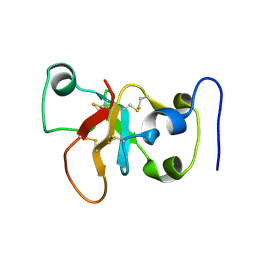 | |
8ON5
 
 | |
7ODF
 
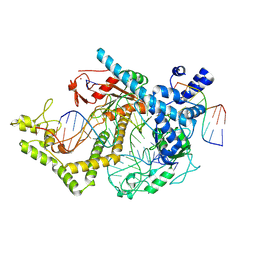 | | Structure of the mini-RNA-guided endonuclease CRISPR-Cas_phi3 | | Descriptor: | Cas_phi3, DNA (5'-D(P*GP*G)-3'), DNA (5'-D(P*GP*TP*AP*AP*TP*TP*CP*AP*G)-3'), ... | | Authors: | Carabias del Rey, A, Fugilsang, A, Temperini, P, Pape, T, Sofos, N, Stella, S, Erledsson, S, Montoya, G. | | Deposit date: | 2021-04-29 | | Release date: | 2021-07-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structure of the mini-RNA-guided endonuclease CRISPR-Cas12j3.
Nat Commun, 12, 2021
|
|
5UJ2
 
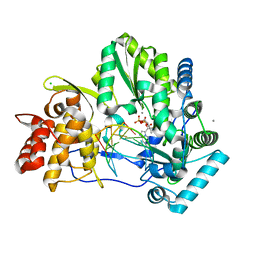 | | Crystal structure of HCV NS5B genotype 2A JFH-1 isolate with S15G E86Q E87Q C223H V321I mutations and Delta8 neta hairpoin loop deletion in complex with GS-639476 (diphsohate version of GS-9813), Mn2+ and symmetrical primer template 5'-AUAAAUUU | | Descriptor: | (1S)-1-(4-aminoimidazo[2,1-f][1,2,4]triazin-7-yl)-1,4-anhydro-2-deoxy-2-fluoro-5-O-[(S)-hydroxy(phosphonooxy)phosphoryl]-2-methyl-D-ribitol, CHLORIDE ION, Genome polyprotein, ... | | Authors: | Edwards, T.E, Fox III, D, Appleby, T.C, Murakami, E, Rey, A, McGrath, M.E. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a 2'-fluoro-2'-C-methyl C-nucleotide HCV polymerase inhibitor and a phosphoramidate prodrug with favorable properties.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
8QBM
 
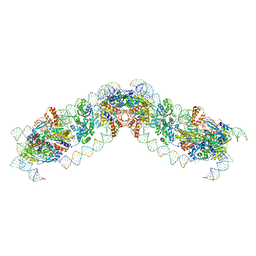 | |
8QBL
 
 | |
8QBK
 
 | |
7L0D
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML188 | | Descriptor: | 3C-like proteinase, N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)furan-2-carboxamide | | Authors: | Lockbaum, G.J, Lee, J.M, Reyes, A.C, Nalivaika, E.A, Ali, A, Yilmaz, N.K, Thompson, P.R, Schiffer, C.A. | | Deposit date: | 2020-12-11 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal Structure of SARS-CoV-2 Main Protease in Complex with the Non-Covalent Inhibitor ML188.
Viruses, 13, 2021
|
|
3TV5
 
 | | Crystal Structure of the humanized carboxyltransferase domain of yeast Acetyl-coA caroxylase in complex with compound 1 | | Descriptor: | (3R)-1'-(9-ANTHRYLCARBONYL)-3-(MORPHOLIN-4-YLCARBONYL)-1,4'-BIPIPERIDINE, Acetyl-CoA carboxylase | | Authors: | Rajamohan, F, Marr, E, Reyes, A, Landro, J.A, Anderson, M.D, Corbett, J.W, Dirico, K.J, Harwood, J.H, Tu, M, Vajdos, F.F. | | Deposit date: | 2011-09-19 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-guided Inhibitor Design for Human Acetyl-coenzyme A Carboxylase by Interspecies Active Site Conversion.
J.Biol.Chem., 286, 2011
|
|
3TVW
 
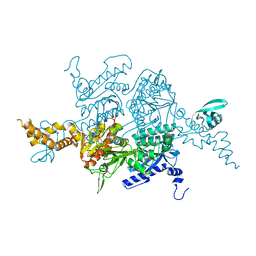 | | Crystal Structure of the humanized carboxyltransferase domain of yeast Acetyl-coA caroxylase in complex with compound 4 | | Descriptor: | Acetyl-CoA carboxylase, [4-(2H-chromen-3-ylmethyl)piperazin-1-yl]-[3-(1H-pyrazol-5-yl)phenyl]methanone | | Authors: | Rajamohan, F, Marr, E, Reyes, A, Landro, J.A, Anderson, M.D, Corbett, J.W, Dirico, K.J, Harwood, J.H, Tu, M, Vajdos, F.F. | | Deposit date: | 2011-09-20 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-guided Inhibitor Design for Human Acetyl-coenzyme A Carboxylase by Interspecies Active Site Conversion.
J.Biol.Chem., 286, 2011
|
|
3TVU
 
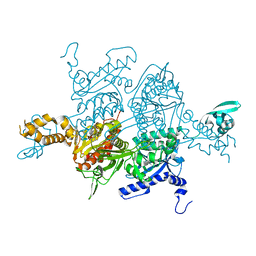 | | Crystal Structure of the humanized carboxyltransferase domain of yeast Acetyl-coA caroxylase in complex with compound 3 | | Descriptor: | 4-({4-[(2-methylquinolin-6-yl)methyl]piperidin-1-yl}carbonyl)-2-phenylquinoline, Acetyl-CoA carboxylase | | Authors: | Rajamohan, F, Marr, E, Reyes, A, Landro, J.A, Anderson, M.D, Corbett, J.W, Dirico, K.J, Harwood, J.H, Tu, M, Vajdos, F.F. | | Deposit date: | 2011-09-20 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-guided Inhibitor Design for Human Acetyl-coenzyme A Carboxylase by Interspecies Active Site Conversion.
J.Biol.Chem., 286, 2011
|
|
3TZ3
 
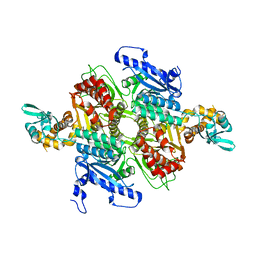 | | Crystal Structure of the humanized carboxyltransferase domain of yeast Acetyl-coA caroxylase in complex with compound 2 | | Descriptor: | 6-{[1-(anthracen-9-ylcarbonyl)piperidin-4-yl]methyl}-2-methylquinoline, Acetyl-CoA carboxylase | | Authors: | Rajamohan, F, Marr, E, Reyes, A, Landro, J.A, Anderson, M.D, Corbett, J.W, Dirico, K.J, Harwood, J.H, Tu, M, Vajdos, F.F. | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-guided Inhibitor Design for Human Acetyl-coenzyme A Carboxylase by Interspecies Active Site Conversion.
J.Biol.Chem., 286, 2011
|
|
6KSN
 
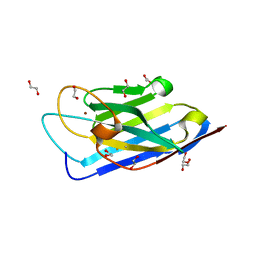 | | Structure of a Zn-bound camelid single domain antibody | | Descriptor: | 1,2-ETHANEDIOL, ICab3, SODIUM ION, ... | | Authors: | Kumar, S, Athreya, A, Penmatsa, A. | | Deposit date: | 2019-08-24 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Isolation and structural characterization of a Zn2+-bound single-domain antibody against NorC, a putative multidrug efflux transporter in bacteria.
J.Biol.Chem., 295, 2020
|
|
6BP2
 
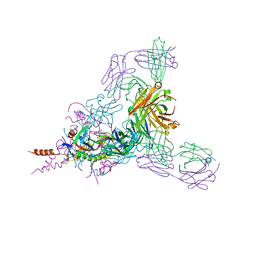 | | Therapeutic human monoclonal antibody MR191 bound to a marburgvirus glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, Envelope glycoprotein GP2, ... | | Authors: | King, L.B, Fusco, M.L, Flyak, A.I, Ilinykh, P.A, Huang, K, Gunn, B, Kirchdoerfer, R.N, Hastie, K.M, Sangha, A.K, Meiler, J, Alter, G, Bukreyev, A, Crowe, J.E.J, Saphire, E.O. | | Deposit date: | 2017-11-21 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.172 Å) | | Cite: | The Marburgvirus-Neutralizing Human Monoclonal Antibody MR191 Targets a Conserved Site to Block Virus Receptor Binding.
Cell Host Microbe, 23, 2018
|
|
5N7E
 
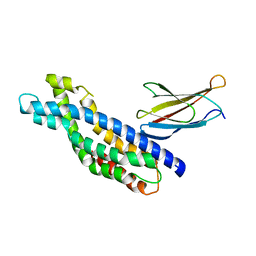 | | Crystal structure of the Dbl-homology domain of Bcr-Abl in complex with monobody Mb(Bcr-DH_4). | | Descriptor: | Breakpoint cluster region protein, Mb(Bcr-DH_4) | | Authors: | Reckel, S, Reynaud, A, Pojer, F, Hantschel, O. | | Deposit date: | 2017-02-20 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Structural and functional dissection of the DH and PH domains of oncogenic Bcr-Abl tyrosine kinase.
Nat Commun, 8, 2017
|
|
5OC7
 
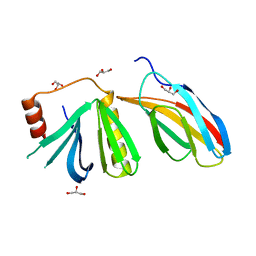 | | Crystal structure of the pleckstrin-homology domain of Bcr-Abl in complex with monobody Mb(Bcr-PH_4). | | Descriptor: | Breakpoint cluster region protein,pleckstrin-homology domain of Bcr-Abl, D-MYO-INOSITOL-4,5-BISPHOSPHATE, GLYCEROL, ... | | Authors: | Reckel, S, Reynaud, A, Pojer, F, Hantschel, O. | | Deposit date: | 2017-06-29 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Structural and functional dissection of the DH and PH domains of oncogenic Bcr-Abl tyrosine kinase.
Nat Commun, 8, 2017
|
|
4EGW
 
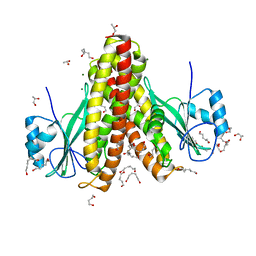 | | The structure of the soluble domain of CorA from Methanocaldococcus jannaschii | | Descriptor: | 1,4-BUTANEDIOL, HEXANE-1,6-DIOL, MAGNESIUM ION, ... | | Authors: | Guskov, A, Nordin, N, Reynaud, A, Engman, H, Lundback, A.-K, Jong, A.J.O, Cornvik, T, Phua, T, Eshaghi, S. | | Deposit date: | 2012-04-02 | | Release date: | 2012-10-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the mechanisms of Mg2+ uptake, transport, and gating by CorA
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4EV6
 
 | | The complete structure of CorA magnesium transporter from Methanocaldococcus jannaschii | | Descriptor: | MAGNESIUM ION, Magnesium transport protein CorA, UNDECYL-MALTOSIDE | | Authors: | Guskov, A, Nordin, N, Reynaud, A, Engman, H, Lundback, A.-K, Jong, A.J.O, Cornvik, T, Phua, T, Eshaghi, S. | | Deposit date: | 2012-04-25 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the mechanisms of Mg2+ uptake, transport, and gating by CorA
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8OKV
 
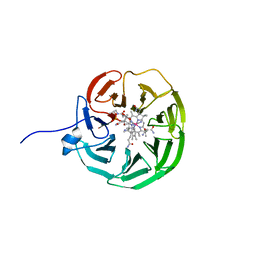 | | lipoprotein BT2095 from Bacteroides thetaiotamicron bound to cyanocobalamin CnCbl | | Descriptor: | CYANOCOBALAMIN, Putative surface layer protein | | Authors: | Javier Abellon-Ruiz, J, van den Berg, B, Jana, K, Kleinekathofer, U, Silale, A, Frey, A.M, Basle, A, Trost, M. | | Deposit date: | 2023-03-29 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | BtuB TonB-dependent transporters and BtuG surface lipoproteins form stable complexes for vitamin B 12 uptake in gut Bacteroides.
Nat Commun, 14, 2023
|
|
6R63
 
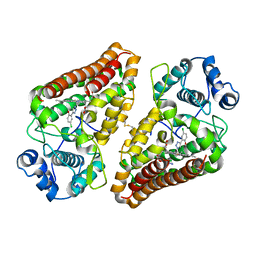 | | Crystal structure of indoleamine 2,3-dioxygenase 1 (IDO1) in complex with ferric heme and MMG-0358 | | Descriptor: | 4-chloranyl-2-(2~{H}-1,2,3-triazol-4-yl)phenol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roehrig, U.F, Reynaud, A, Pojer, F, Michielin, O, Zoete, V. | | Deposit date: | 2019-03-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Inhibition Mechanisms of Indoleamine 2,3-Dioxygenase 1 (IDO1).
J.Med.Chem., 62, 2019
|
|
7AH4
 
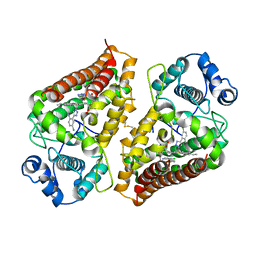 | | Crystal structure of indoleamine 2,3-dioxygenase 1 (IDO1) in complex with ferric heme and MMG-0363 | | Descriptor: | 4-chloranyl-2-(2~{H}-1,2,3-triazol-4-yl)aniline, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roehrig, U.F, Reynaud, A, Pojer, F, Michielin, O, Zoete, V. | | Deposit date: | 2020-09-24 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Azole-Based Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AH6
 
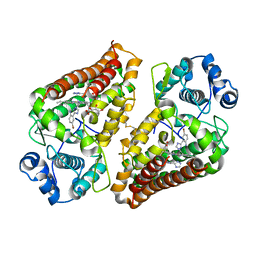 | | Crystal structure of indoleamine 2,3-dioxygenase 1 (IDO1) in complex with ferric heme and MMG-0752 | | Descriptor: | 4-bromanyl-2-(4~{H}-1,2,4-triazol-3-yl)aniline, GLYCEROL, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Roehrig, U.F, Reynaud, A, Pojer, F, Michielin, O, Zoete, V. | | Deposit date: | 2020-09-24 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Azole-Based Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AH5
 
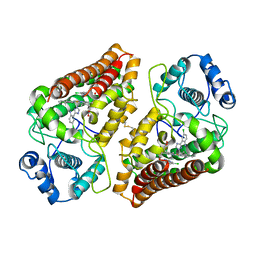 | | Crystal structure of indoleamine 2,3-dioxygenase 1 (IDO1) in complex with ferric heme and MMG-0706 | | Descriptor: | 4-chloranyl-2-(1~{H}-1,2,4-triazol-5-yl)aniline, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roehrig, U.F, Reynaud, A, Pojer, F, Michielin, O, Zoete, V. | | Deposit date: | 2020-09-24 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Azole-Based Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7D5Q
 
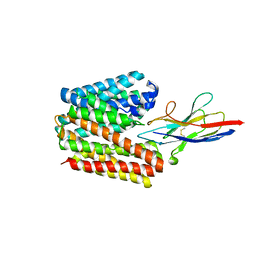 | | Structure of NorC transporter (K398A mutant) in an outward-open conformation in complex with a single-chain Indian camelid antibody | | Descriptor: | Drug transporter, putative, ICab, ... | | Authors: | Kumar, S, Athreya, A, Penmatsa, A. | | Deposit date: | 2020-09-27 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis of inhibition of a transporter from Staphylococcus aureus, NorC, through a single-domain camelid antibody.
Commun Biol, 4, 2021
|
|
7D5P
 
 | | Structure of NorC transporter in an outward-open conformation in complex with a single-chain Indian camelid antibody | | Descriptor: | Drug transporter, putative, ICab, ... | | Authors: | Kumar, S, Athreya, A, Penmatsa, A. | | Deposit date: | 2020-09-27 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural basis of inhibition of a transporter from Staphylococcus aureus, NorC, through a single-domain camelid antibody.
Commun Biol, 4, 2021
|
|
