1R3N
 
 | | Crystal structure of beta-alanine synthase from Saccharomyces kluyveri | | Descriptor: | BETA-AMINO ISOBUTYRATE, ZINC ION, beta-alanine synthase | | Authors: | Lundgren, S, Gojkovic, Z, Piskur, J, Dobritzsch, D. | | Deposit date: | 2003-10-02 | | Release date: | 2003-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Yeast beta-Alanine Synthase Shares a Structural Scaffold and Origin with Dizinc-dependent Exopeptidases
J.Biol.Chem., 278, 2003
|
|
1E8V
 
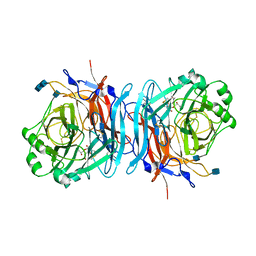 | | Structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Crennell, S, Takimoto, T, Portner, A, Taylor, G. | | Deposit date: | 2000-10-01 | | Release date: | 2001-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Multifunctional Paramyxovirus Hemagglutinin-Neuraminidase
Nat.Struct.Biol., 7, 2000
|
|
7BPU
 
 | |
5GUF
 
 | |
6ELJ
 
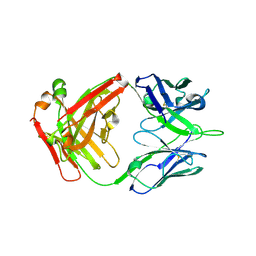 | | FAB Fragment. AbVance: Increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery | | Descriptor: | fAB heavy chain, fAB light chain | | Authors: | Benz, J, Weigand, S, Dengl, S, Schlothauer, T, Auer, J, Ehler, A, Kettenberger, H, Lorenz, S, Hirschheydt, T, Georges, G. | | Deposit date: | 2017-09-29 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AbVance: increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery
To Be Published
|
|
6ELE
 
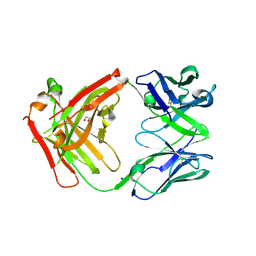 | | FAB Fragment. AbVance: Increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, fAB heavy chain, ... | | Authors: | Benz, J, Weigand, S, Dengl, S, Schlothauer, T, Auer, J, Ehler, A, Kettenberger, H, Lorenz, S, Hirschheydt, T, Georges, G. | | Deposit date: | 2017-09-28 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | AbVance: increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery
To Be Published
|
|
6ELL
 
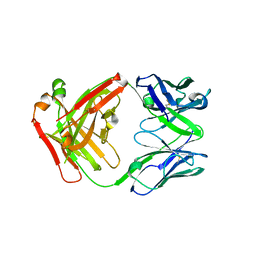 | | FAB Fragment. AbVance: Increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery | | Descriptor: | fAB heavy chain, fAB light chain | | Authors: | Benz, J, Weigand, S, Dengl, S, Schlothauer, T, Auer, J, Ehler, A, Kettenberger, H, Lorenz, S, Hirschheydt, T, Georges, G. | | Deposit date: | 2017-09-29 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AbVance: increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery
To Be Published
|
|
6EMJ
 
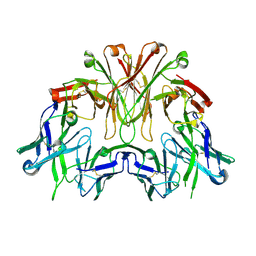 | | FAB Fragment. AbVance: Increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery | | Descriptor: | SODIUM ION, fAB heavy chain, fAb light chain | | Authors: | Benz, J, Weigand, S, Dengl, S, Schlothauer, T, Auer, J, Ehler, A, Kettenberger, H, Lorenz, S, Hirschheydt, T, Georges, G. | | Deposit date: | 2017-10-02 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AbVance: increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery
To Be Published
|
|
6ENU
 
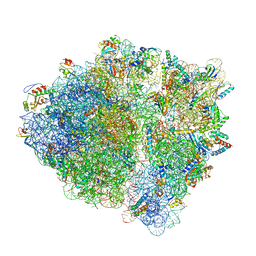 | | Polyproline-stalled ribosome in the presence of elongation-factor P (EF-P) | | Descriptor: | 16S ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Huter, P, Arenz, S, Wilson, D.N. | | Deposit date: | 2017-10-06 | | Release date: | 2017-11-22 | | Last modified: | 2018-01-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis for Polyproline-Mediated Ribosome Stalling and Rescue by the Translation Elongation Factor EF-P.
Mol. Cell, 68, 2017
|
|
6ENF
 
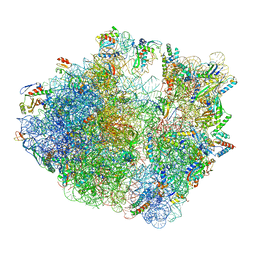 | | Cryo-EM structure of a polyproline-stalled ribosome in the absence of EF-P | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Huter, P, Arenz, S, Wilson, D.N. | | Deposit date: | 2017-10-04 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Basis for Polyproline-Mediated Ribosome Stalling and Rescue by the Translation Elongation Factor EF-P.
Mol. Cell, 68, 2017
|
|
3ZLT
 
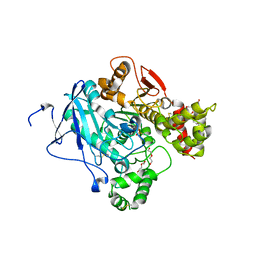 | | Crystal structure of acetylcholinesterase in complex with RVX | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Artursson, E, Andersson, P.O, Akfur, C, Linusson, A, Borjegren, S, Ekstrom, F. | | Deposit date: | 2013-02-04 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalytic-Site Conformational Equilibrium in Nerve-Agent Adducts of Acetylcholinesterase; Possible Implications for the Hi-6 Antidote Substrate Specificity.
Biochem.Pharmacol., 85, 2013
|
|
3ZLU
 
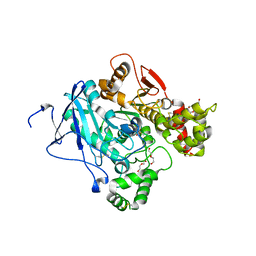 | | Crystal structure of mouse acetylcholinesterase in complex with cyclosarin | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, ACETYLCHOLINESTERASE | | Authors: | Artursson, E, Andersson, P.O, Akfur, C, Linusson, A, Borjegren, S, Ekstrom, F. | | Deposit date: | 2013-02-04 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalytic-Site Conformational Equilibrium in Nerve-Agent Adducts of Acetylcholinesterase; Possible Implications for the Hi-6 Antidote Substrate Specificity.
Biochem.Pharmacol., 85, 2013
|
|
9BOG
 
 | | Structural basis for adhesin secretion by the outer-membrane usher in type 1 pili | | Descriptor: | Outer membrane usher protein FimD, Protein FimF, Type 1 fimbria chaperone FimC, ... | | Authors: | Bitter, R.M, Zimmerman, M, Hultgren, S, Yuan, P. | | Deposit date: | 2024-05-03 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural basis for adhesin secretion by the outer-membrane usher in type 1 pili.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6QH3
 
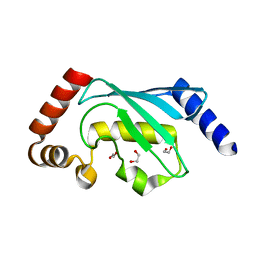 | |
6QHK
 
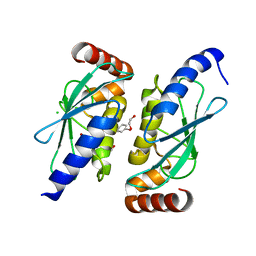 | |
6WTH
 
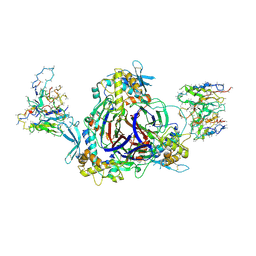 | | Full-length human ENaC ECD | | Descriptor: | 10D4 Fab, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Posert, R, Baconguis, I, Noreng, S, Bharadwaj, A, Houser, A. | | Deposit date: | 2020-05-02 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Molecular principles of assembly, activation, and inhibition in epithelial sodium channel.
Elife, 9, 2020
|
|
6R4O
 
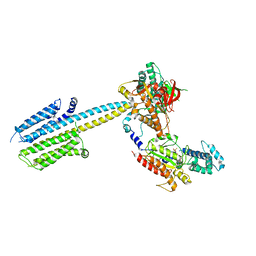 | | Structure of a truncated adenylyl cyclase bound to MANT-GTP, forskolin and an activated stimulatory Galphas protein | | Descriptor: | 3'-O-(N-METHYLANTHRANILOYL)-GUANOSINE-5'-TRIPHOSPHATE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Adenylate cyclase 9, ... | | Authors: | Qi, C, Sorrentino, S, Medalia, O, Korkhov, V.M. | | Deposit date: | 2019-03-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structure of a membrane adenylyl cyclase bound to an activated stimulatory G protein.
Science, 364, 2019
|
|
6XZ1
 
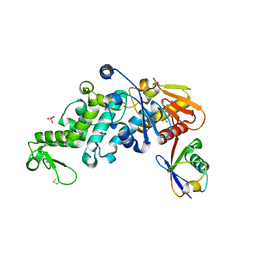 | | Conjugate of the HECT domain of HUWE1 with ubiquitin | | Descriptor: | HECT, UBA and WWE domain containing 1, isoform CRA_a, ... | | Authors: | Liu, B, Seenivasan, A, Nair, R, Chen, D, Lowe, E.D, Lorenz, S. | | Deposit date: | 2020-01-31 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reconstitution and Structural Analysis of a HECT Ligase-Ubiquitin Complex via an Activity-Based Probe.
Acs Chem.Biol., 16, 2021
|
|
7AZW
 
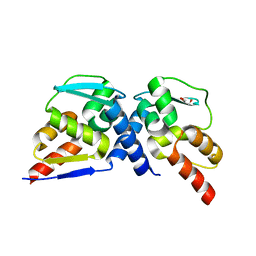 | | Crystal structure of the MIZ1-BTB-domain | | Descriptor: | GLYCEROL, Zinc finger and BTB domain-containing protein 17 isoform X1 | | Authors: | Orth, B, Sander, B, Diederichs, K, Lorenz, S. | | Deposit date: | 2020-11-17 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of an atypical interaction site in the BTB domain of the MYC-interacting zinc-finger protein 1.
Structure, 29, 2021
|
|
7AZX
 
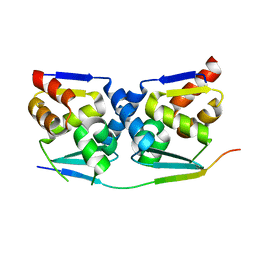 | | Crystal structure of the MIZ1-BTB-domain in complex with a HUWE1-derived peptide | | Descriptor: | E3 ubiquitin-protein ligase HUWE1, Zinc finger and BTB domain-containing protein 17 isoform X1 | | Authors: | Orth, B, Sander, B, Diederichs, K, Lorenz, S. | | Deposit date: | 2020-11-17 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of an atypical interaction site in the BTB domain of the MYC-interacting zinc-finger protein 1.
Structure, 29, 2021
|
|
7AHF
 
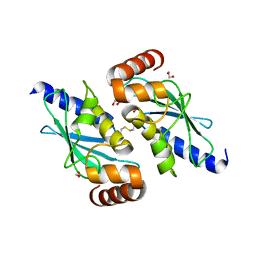 | |
8Q0N
 
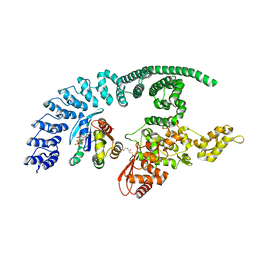 | | HACE1 in complex with RAC1 Q61L | | Descriptor: | E3 ubiquitin-protein ligase HACE1, GUANOSINE-5'-TRIPHOSPHATE, Ras-related C3 botulinum toxin substrate 1, ... | | Authors: | Wolter, M, Duering, J, Dienemann, C, Lorenz, S. | | Deposit date: | 2023-07-28 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mechanisms of autoinhibition and substrate recognition by the ubiquitin ligase HACE1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8PWL
 
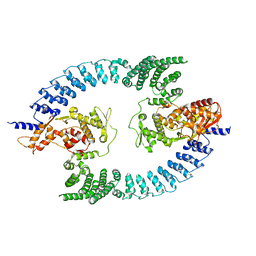 | |
5NJT
 
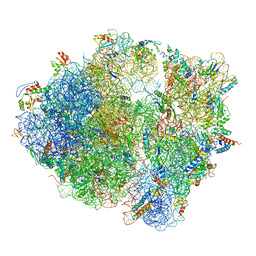 | | Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Beckert, B, Abdelshahid, M, Schaefer, H, Steinchen, W, Arenz, S, Berninghausen, O, Beckmann, R, Bange, G, Turgay, K, Wilson, D.N. | | Deposit date: | 2017-03-29 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization.
EMBO J., 36, 2017
|
|
5MGP
 
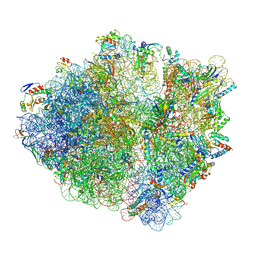 | | Structural basis for ArfA-RF2 mediated translation termination on stop-codon lacking mRNAs | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Huter, P, Mueller, C, Beckert, B, Arenz, S, Berninghausen, O, Beckmann, R, Wilson, N.D. | | Deposit date: | 2016-11-21 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for ArfA-RF2-mediated translation termination on mRNAs lacking stop codons.
Nature, 541, 2017
|
|
