8ES4
 
 | |
7SAG
 
 | |
7QCM
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3-methoxy-4-hydroxy-acetophenone)thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3-metoxy-4-hydroxy-acetophenone)thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCJ
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2,4-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(2,4-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCG
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2-pyrrolidyl)-3,4,5-trihydroxybenzoylhydrazone | | Descriptor: | 3,4,5-tris(oxidanyl)-N-[(E)-1H-pyrrol-2-ylmethylideneamino]benzamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-23 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCK
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2,5-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(2,5-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCI
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3,4-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3,4-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCH
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3,5-dimethoxy-4-hydroxybenzyliden)thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3,5-dimetoxy-4-hydroxybenzyliden)thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
8ADN
 
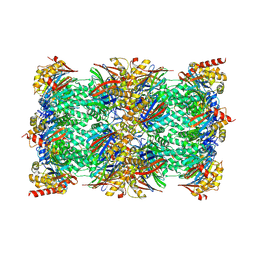 | | Vairimorpha necatrix 20S proteasome from spores | | Descriptor: | Proteasome Inhibitor 31-Like, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Jespersen, N, Ehrenbolger, K, Winiger, R, Svedberg, D, Vossbrinck, C.R, Barandun, J. | | Deposit date: | 2022-07-08 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structure of the reduced microsporidian proteasome bound by PI31-like peptides in dormant spores.
Nat Commun, 13, 2022
|
|
8AVD
 
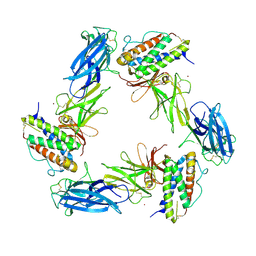 | | Cryo-EM structure for a 3:3 complex between mouse leptin and the mouse LEP-R ectodomain (local refinement) | | Descriptor: | Leptin, Leptin receptor, NICKEL (II) ION | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AVF
 
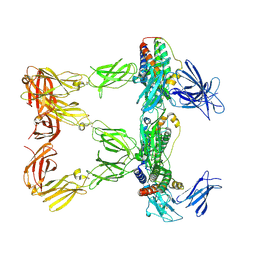 | | Human leptin in complex with the human LEP-R ectodomain fused to a C-terminal trimeric isoleucine GCN4 zipper (closed 3:3 model) | | Descriptor: | Leptin, Leptin receptor | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (6.45 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AVE
 
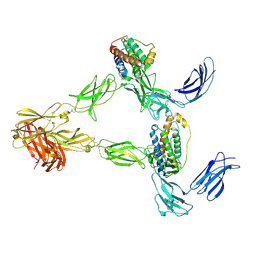 | | Human leptin in complex with the human LEP-R ectodomain fused to a C-terminal trimeric isoleucine GCN4 zipper (2:2 model) | | Descriptor: | Leptin, Leptin receptor | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.62 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AVO
 
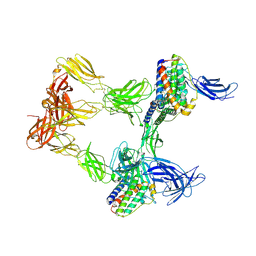 | | Human leptin in complex with the human LEP-R ectodomain fused to a C-terminal trimeric isoleucine GCN4 zipper (open 3:3 model). | | Descriptor: | Leptin, Leptin receptor | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.84 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AVC
 
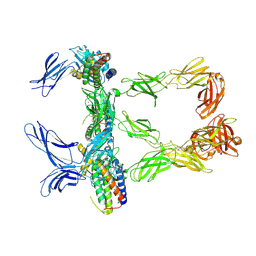 | | Mouse leptin:LEP-R complex cryoEM structure (3:3 model) | | Descriptor: | Leptin, Leptin receptor, NICKEL (II) ION | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4PBU
 
 | | Serial Time-resolved crystallography of Photosystem II using a femtosecond X-ray laser The S1 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Kupitz, C, Basu, S, Grotjohann, I, Fromme, R, Zatsepin, N, Rendek, K.N, Hunter, M, Shoeman, R.L, White, T.A, Wang, D, James, D, Yang, J.H, Cobb, D.E, Reeder, B, Sierra, R.G, Liu, H, Barty, A, Aquila, A, Deponte, D, Kirian, R.A, Bari, S, Bergkamp, J.J, Beyerlein, K, Bogan, M.J, Caleman, C, Chao, T.-C, Conrad, C.E, Davis, K.M, Fleckenstein, H, Galli, L, Hau-Riege, S.P, Kassemeyer, S, Laksmono, H, Liang, M, Lomb, L, Marchesini, S, Martin, A.V, Messerschmidt, M, Milathianaki, D, Nass, K, Ros, A, Roy-Chowdhury, S, Schmidt, K, Seibert, M, Steinbrener, J, Stellato, F, Yan, L, Yoon, C, Moore, T.A, Moore, A.L, Pushkar, Y, Williams, G.J, Boutet, S, Doak, R.B, Weierstall, U, Frank, M, Chapman, H.N, Spence, J.C.H, Fromme, P. | | Deposit date: | 2014-04-13 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser.
Nature, 513, 2014
|
|
8V2V
 
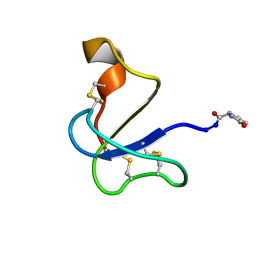 | |
7PXZ
 
 | | Reduced form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-08 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
7PZQ
 
 | | Oxidized form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
6WQJ
 
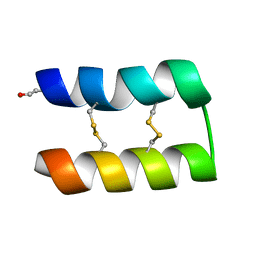 | |
6WQL
 
 | |
7RC8
 
 | | Solution NMR Structure of [D-Ala19]Crp4 | | Descriptor: | Alpha-defensin 4 | | Authors: | Conibear, A.C, Rosengren, K.J. | | Deposit date: | 2021-07-07 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | A conserved beta-bulge glycine residue facilitates folding and increases stability of the mouse alpha-defensin cryptdin-4
Peptide Science, 114, 2022
|
|
7M25
 
 | |
7M2A
 
 | |
6CEI
 
 | |
7N1Z
 
 | | NMR structure of native PnIA | | Descriptor: | Alpha-conotoxin PnIA | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
