5FSS
 
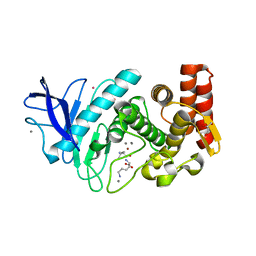 | | Structure of thermolysin prepared by the 'soak-and-freeze' method under 40 bar of krypton pressure | | Descriptor: | CALCIUM ION, GLYCEROL, KRYPTON, ... | | Authors: | Lafumat, B, Mueller-Dieckmann, C, Colloc'h, N, Prange, T, Royant, A, van der Linden, P, Carpentier, P. | | Deposit date: | 2016-01-07 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Gas-Sensitive Biological Crystals Processed in Pressurized Oxygen and Krypton Atmospheres: Deciphering Gas Channels in Proteins Using a Novel `Soak-and-Freeze' Methodology.
J.Appl.Crystallogr., 49, 2016
|
|
1OJT
 
 | |
5FXJ
 
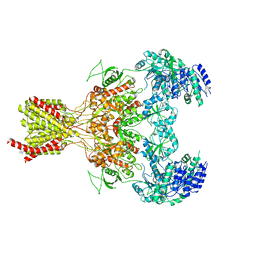 | | GluN1b-GluN2B NMDA receptor structure-Class X | | Descriptor: | GLUTAMATE RECEPTOR IONOTROPIC, NMDA 1, NMDA 2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa H, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.25 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5FSP
 
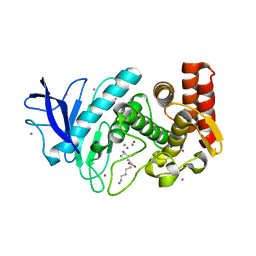 | | Structure of thermolysin prepared by the 'soak-and-freeze' method under 100 bar of krypton pressure | | Descriptor: | CALCIUM ION, KRYPTON, LYSINE, ... | | Authors: | Lafumat, B, Mueller-Dieckmann, C, Colloc'h, N, Prange, T, Royant, A, van der Linden, p, Carpentier, P. | | Deposit date: | 2016-01-06 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Gas-Sensitive Biological Crystals Processed in Pressurized Oxygen and Krypton Atmospheres: Deciphering Gas Channels in Proteins Using a Novel `Soak-and-Freeze' Methodology.
J.Appl.Crystallogr., 49, 2016
|
|
5FXI
 
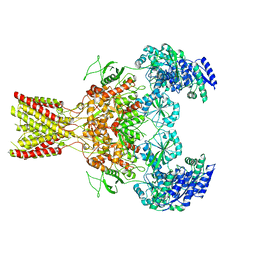 | | GluN1b-GluN2B NMDA receptor structure in non-active-2 conformation | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5FST
 
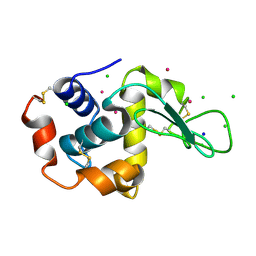 | | Structure of lysozyme prepared by the 'soak-and-freeze' method under 100 bar of krypton pressure | | Descriptor: | CHLORIDE ION, KRYPTON, LYSOZYME C, ... | | Authors: | Lafumat, B, Mueller-Dieckmann, C, Colloc'h, N, Prange, T, Royant, A, van der Linden, P, Carpentier, P. | | Deposit date: | 2016-01-07 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Gas-Sensitive Biological Crystals Processed in Pressurized Oxygen and Krypton Atmospheres: Deciphering Gas Channels in Proteins Using a Novel `Soak-and-Freeze' Methodology.
J.Appl.Crystallogr., 49, 2016
|
|
5FXK
 
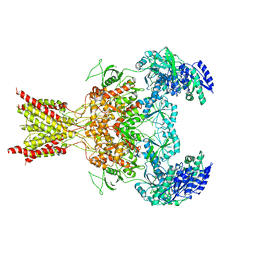 | | GluN1b-GluN2B NMDA receptor structure-Class Y | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5FXG
 
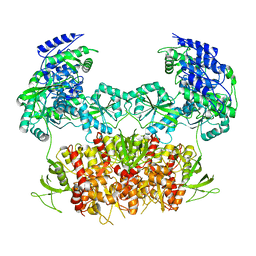 | | GLUN1B-GLUN2B NMDA RECEPTOR IN ACTIVE CONFORMATION | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
1M7E
 
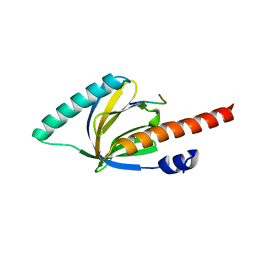 | | Crystal structure of the phosphotyrosine binding domain(PTB) of mouse Disabled 2(Dab2):implications for Reeling signaling | | Descriptor: | Disabled homolog 2, NGYENPTYK peptide | | Authors: | Yun, M, Keshvara, L, Park, C.-G, Zhang, Y.-M, Dickerson, J.B, Zheng, J, Rock, C.O, Curran, T, Park, H.-W. | | Deposit date: | 2002-07-19 | | Release date: | 2003-08-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of the Dab homology domains of mouse disabled 1 and 2
J.Biol.Chem., 278, 2003
|
|
1WRR
 
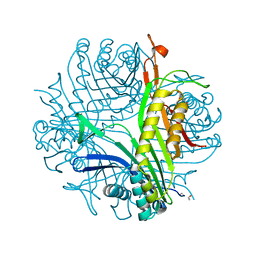 | | Urate oxidase from aspergillus flavus complexed with 5-amino 6-nitro uracil | | Descriptor: | 5-AMINO-6-NITROPYRIMIDINE-2,4(1H,3H)-DIONE, Uricase | | Authors: | Retailleau, P, Colloc'h, N, Vivares, D, Bonnete, F, Castro, B, El Hajji, M, Prange, T. | | Deposit date: | 2004-10-27 | | Release date: | 2005-03-22 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Urate oxidase from Aspergillus flavus: new crystal-packing contacts in relation to the content of the active site.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1WS3
 
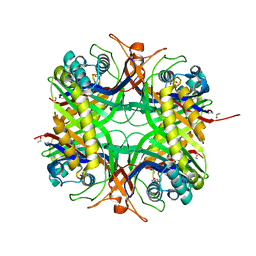 | | Urate oxidase from aspergillus flavus complexed with uracil | | Descriptor: | URACIL, Uricase | | Authors: | Retailleau, P, Colloc'h, N, Vivares, D, Bonnete, F, Castro, B, El Hajji, M, Prange, T. | | Deposit date: | 2004-10-29 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Urate oxidase from Aspergillus flavus: new crystal-packing contacts in relation to the content of the active site.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ICG
 
 | | STRUCTURE OF THE HYBRID RNA/DNA R-GCUUCGGC-D[F]U IN PRESENCE OF IR(NH3)6+++ | | Descriptor: | 5'-R(*GP*CP*UP*UP*CP*GP*GP*C)-D(P*(UFP))-3', CHLORIDE ION, IRIDIUM HEXAMMINE ION | | Authors: | Cruse, W, Saludjian, P, Neuman, A, Prange, T. | | Deposit date: | 2001-03-31 | | Release date: | 2001-04-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Destabilizing effect of a fluorouracil extra base in a hybrid RNA duplex compared with bromo and chloro analogues.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7C06
 
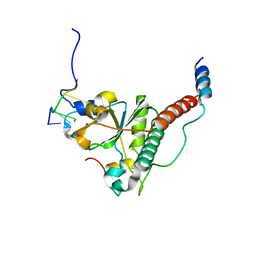 | | Crystal structure of yeast U2AF1 complex bound to 3' splice site RNA, 5'-UAGGU. | | Descriptor: | RNA (5'-R(*U*UP*AP*GP*GP*U)-3'), Splicing factor U2AF 23 kDa subunit, Splicing factor U2AF 59 kDa subunit, ... | | Authors: | Yoshida, H, Park, S.Y, Urano, T, Obayashi, E. | | Deposit date: | 2020-04-30 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Elucidation of the aberrant 3' splice site selection by cancer-associated mutations on the U2AF1.
Nat Commun, 11, 2020
|
|
7C08
 
 | | Crystal structure of S34Y mutant of yeast U2AF1 complex bound to 3' splice site RNA, 5'-UAGGU. | | Descriptor: | RNA (5'-R(*U*UP*AP*GP*GP*U)-3'), Splicing factor U2AF 23 kDa subunit, Splicing factor U2AF 59 kDa subunit, ... | | Authors: | Yoshida, H, Park, S.Y, Urano, T, Obayashi, E. | | Deposit date: | 2020-04-30 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Elucidation of the aberrant 3' splice site selection by cancer-associated mutations on the U2AF1.
Nat Commun, 11, 2020
|
|
1IHA
 
 | | Structure of the Hybrid RNA/DNA R-GCUUCGGC-D[BR]U in Presence of RH(NH3)6+++ | | Descriptor: | 5'-R(*GP*CP*UP*UP*CP*GP*GP*C)-D(P*(BRU))-3', CHLORIDE ION, RHODIUM HEXAMINE ION | | Authors: | Cruse, W.B, Saludjian, P, Neuman, A, Prange, T. | | Deposit date: | 2001-04-19 | | Release date: | 2001-05-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Destabilizing effect of a fluorouracil extra base in a hybrid RNA duplex compared with bromo and chloro analogues.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IDW
 
 | | STRUCTURE OF THE HYBRID RNA/DNA R-GCUUCGGC-D[CL]U IN PRESENCE OF RH(NH3)6+++ | | Descriptor: | 5'-R(*GP*CP*UP*UP*CP*GP*GP*C)-D(P*(UCL))-3', CHLORIDE ION, RHODIUM HEXAMINE ION | | Authors: | Cruse, W, Saludjian, P, Neuman, A, Prange, T. | | Deposit date: | 2001-04-05 | | Release date: | 2001-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Destabilizing effect of a fluorouracil extra base in a hybrid RNA duplex compared with bromo and chloro analogues
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1ISG
 
 | | Crystal Structure Analysis of BST-1/CD157 with ATPgammaS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, bone marrow stromal cell antigen 1 | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
1ISM
 
 | | Crystal Structure Analysis of BST-1/CD157 complexed with nicotinamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NICOTINAMIDE, bone marrow stromal cell antigen 1 | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
6DHP
 
 | | RT XFEL structure of the three-flash state of Photosystem II (3F, S0-rich) at 2.04 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
1WS2
 
 | | urate oxidase from aspergillus flavus complexed with 5,6-diaminouracil | | Descriptor: | 5,6-DIAMINOPYRIMIDINE-2,4(1H,3H)-DIONE, Uricase | | Authors: | Retailleau, P, Colloc'h, N, Vivares, D, Bonnete, F, Castro, B, El Hajji, M, Prange, T. | | Deposit date: | 2004-10-29 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Urate oxidase from Aspergillus flavus: new crystal-packing contacts in relation to the content of the active site.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ISJ
 
 | | Crystal Structure Analysis of BST-1/CD157 complexed with NMN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, bone marrow stromal cell antigen 1 | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
1ISI
 
 | | Crystal Structure Analysis of BST-1/CD157 complexed with ethenoNAD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NICOTINAMIDE, [[(2R,3S,4R,5R)-3,4-dihydroxy-5-(9H-imidazo[2,1-f]purin-6-ium-3-yl)oxolan-2-yl]methoxy-oxidanidyl-phosphoryl] [(2R,3S,4R,5R)-3,4-dihydroxy-5-oxidanidyl-oxolan-2-yl]methyl phosphate, ... | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
7EST
 
 | |
376D
 
 | | A ZIPPER-LIKE DNA DUPLEX D(GCGAAAGCT) | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*(CBR)P*GP*AP*AP*AP*GP*CP*T)-3') | | Authors: | Cruse, W.B.T, Shepard, W, Prange, T, delalFortelle, E, Fourme, R. | | Deposit date: | 1998-01-22 | | Release date: | 1999-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A zipper-like duplex in DNA: the crystal structure of d(GCGAAAGCT) at 2.1 A resolution.
Structure, 6, 1998
|
|
1VT7
 
 | |
