6EBM
 
 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs, transmembrane domain of subunit alpha | | 分子名称: | Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera | | 著者 | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | 登録日 | 2018-08-06 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
2G5N
 
 | |
2G5V
 
 | |
2GH1
 
 | | Crystal Structure of the putative SAM-dependent methyltransferase BC2162 from Bacillus cereus, Northeast Structural Genomics Target BcR20. | | 分子名称: | ACETATE ION, GLYCEROL, Methyltransferase | | 著者 | Forouhar, F, Neely, H, Jayaraman, S, Ciao, M, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-03-24 | | 公開日 | 2006-04-04 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of the putative SAM-dependent methyltransferase BC2162 from Bacillus cereus, Northeast Structural Genomics Target BcR20.
To be Published
|
|
2GAN
 
 | | Crystal Structure of a Putative Acetyltransferase from Pyrococcus horikoshii, Northeast Structural Genomics Target JR32. | | 分子名称: | 1,2-ETHANEDIOL, 182aa long hypothetical protein, SULFATE ION | | 著者 | Forouhar, F, Abashidze, M, Jayaraman, S, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-03-09 | | 公開日 | 2006-03-21 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of a Putative Acetyltransferase from Pyrococcus horikoshii, Northeast Structural Genomics Target JR32.
To be Published
|
|
2G9D
 
 | | Crystal Structure of Succinylglutamate desuccinylase from Vibrio cholerae, Northeast Structural Genomics Target VcR20 | | 分子名称: | Succinylglutamate desuccinylase | | 著者 | Zhou, W, Jayaraman, S, Forouhar, F, Conover, K, Rong, X, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-03-06 | | 公開日 | 2006-04-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal Structure of Succinylglutamate desuccinylase from Vibrio cholerae, Northeast Structural Genomics Target VcR20.
To be Published
|
|
2GSV
 
 | | X-Ray Crystal Structure of Protein YvfG from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR478. | | 分子名称: | Hypothetical protein yvfG, SULFATE ION | | 著者 | Forouhar, F, Su, M, Jayaraman, S, Wang, D, Fang, Y, Cunningham, K, Conover, K, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-04-26 | | 公開日 | 2006-05-09 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of the Hypothetical Protein YvfG from Bacillus subtilis, Northeast Structural Genomics Target SR478
To be Published
|
|
2GF4
 
 | | Crystal structure of Vng1086c from Halobacterium salinarium (Halobacterium halobium). Northeast Structural Genomics Target HsR14 | | 分子名称: | ACETATE ION, CALCIUM ION, Protein Vng1086c | | 著者 | Benach, J, Zhou, W, Jayaraman, S, Forouhar, F.F, Janjua, H, Xiao, R, Ma, L.-C, Cunningham, K, Wang, D, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-03-21 | | 公開日 | 2006-04-18 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Crystal structure of Vng1086c from Halobacterium salinarium (Halobacterium halobium). Northeast Structural Genomics Target HsR14
To be Published
|
|
8UNH
 
 | | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp | | 分子名称: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Sliding clamp, ... | | 著者 | Huang, Y, Marcus, K, Subramanian, S, Gee, L.C, Gorday, K, Ghaffari-Kashani, S, Luo, X, Zhang, L, O'Donnell, M, Subramanian, S, Kuriyan, J. | | 登録日 | 2023-10-19 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.21 Å) | | 主引用文献 | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8UNF
 
 | | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp and DNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Sliding clamp, ... | | 著者 | Huang, Y, Marcus, K, Subramanian, S, Gee, L.C, Gorday, K, Ghaffari-Kashani, S, Luo, X, Zhang, L, O'Donnell, M, Subramanian, S, Kuriyan, J. | | 登録日 | 2023-10-18 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5JYO
 
 | | Allosteric inhibition of Kidney Isoform of Glutaminase | | 分子名称: | 2-(pyridin-2-yl)-N-(5-{4-[6-({[3-(trifluoromethoxy)phenyl]acetyl}amino)pyridazin-3-yl]butyl}-1,3,4-thiadiazol-2-yl)acetamide, Glutaminase kidney isoform, mitochondrial | | 著者 | Sivaraman, J, Jayaraman, S. | | 登録日 | 2016-05-15 | | 公開日 | 2016-08-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.098 Å) | | 主引用文献 | Structural basis for exploring the allosteric inhibition of human kidney type glutaminase.
Oncotarget, 7, 2016
|
|
6R25
 
 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 3 | | 分子名称: | DNA (147-MER), FLAVIN-ADENINE DINUCLEOTIDE, H2B, ... | | 著者 | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | 登録日 | 2019-03-15 | | 公開日 | 2019-04-24 | | 実験手法 | ELECTRON MICROSCOPY (4.61 Å) | | 主引用文献 | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
8VKP
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | 登録日 | 2024-01-09 | | 公開日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
8VKK
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | 登録日 | 2024-01-09 | | 公開日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.81 Å) | | 主引用文献 | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
8VKM
 
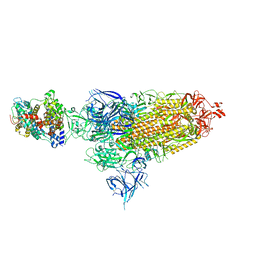 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | 登録日 | 2024-01-09 | | 公開日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
8VKO
 
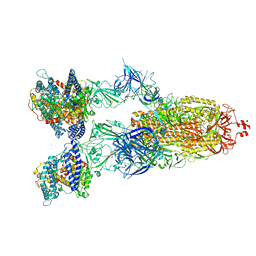 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | 登録日 | 2024-01-09 | | 公開日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
8VKN
 
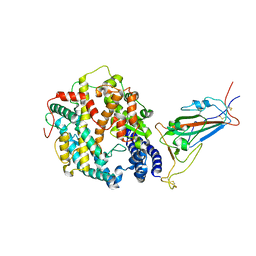 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (focused refinement of RBD and mouse ACE2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | 登録日 | 2024-01-09 | | 公開日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
8VKL
 
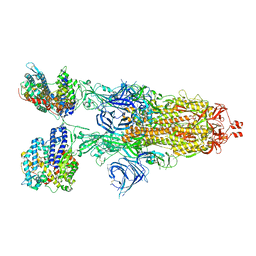 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | 登録日 | 2024-01-09 | | 公開日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
6VYF
 
 | | Cryo-EM structure of Plasmodium vivax hexokinase (Open state) | | 分子名称: | Phosphotransferase | | 著者 | Srivastava, S.S, Darling, J.E, Suryadi, J, Morris, J.C, Drew, M.E, Subramaniam, S. | | 登録日 | 2020-02-26 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Plasmodium vivax and human hexokinases share similar active sites but display distinct quaternary architectures
Iucrj, 7, 2020
|
|
6VYG
 
 | | Cryo-EM structure of Plasmodium vivax hexokinase (Closed state) | | 分子名称: | Phosphotransferase | | 著者 | Srivastava, S.S, Darling, J.E, Suryadi, J, Morris, J.C, Drew, M.E, Subramaniam, S. | | 登録日 | 2020-02-26 | | 公開日 | 2020-05-06 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Plasmodium vivax and human hexokinases share similar active sites but display distinct quaternary architectures
Iucrj, 7, 2020
|
|
6R1T
 
 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 1, free nuclesome | | 分子名称: | DNA (147-MER), HISTONE H2A, Histone H2A, ... | | 著者 | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | 登録日 | 2019-03-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.02 Å) | | 主引用文献 | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6R1U
 
 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 2 | | 分子名称: | DNA (147-MER), FLAVIN-ADENINE DINUCLEOTIDE, Histone H2A, ... | | 著者 | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | 登録日 | 2019-03-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.36 Å) | | 主引用文献 | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6OSI
 
 | | Unmodified tRNA(Pro) bound to Thermus thermophilus 70S (near cognate) | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Hoffer, E.D, Subaramanian, S, Hong, S, Maehigashi, T, Dunham, C.M. | | 登録日 | 2019-05-01 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (4.14 Å) | | 主引用文献 | Structural insights into mRNA reading frame regulation by tRNA modification and slippery codon-anticodon pairing.
Elife, 9, 2020
|
|
8SEV
 
 | | Cryo-EM Structure of RyR1 + ATP-gamma-S (Local Refinement of TMD) | | 分子名称: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Ryanodine receptor 1, ZINC ION | | 著者 | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | 登録日 | 2023-04-10 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEO
 
 | | Cryo-EM Structure of RyR1 + ATP-gamma-S | | 分子名称: | Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Ryanodine receptor 1, ... | | 著者 | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | 登録日 | 2023-04-10 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.92 Å) | | 主引用文献 | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
