2P91
 
 | | Crystal structure of Enoyl-[acyl-carrier-protein] reductase (NADH) from Aquifex aeolicus VF5 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Chen, L, Li, Y, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhao, M, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-23 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Enoyl-[acyl-carrier-protein] reductase (NADH) from Aquifex aeolicus VF5
To be Published
|
|
1ONL
 
 | | Crystal structure of Thermus thermophilus HB8 H-protein of the glycine cleavage system | | Descriptor: | glycine cleavage system H protein | | Authors: | Nakai, T, Ishijima, J, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-28 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Thermus thermophilus HB8 H-protein of the glycine-cleavage system, resolved by a six-dimensional molecular-replacement method.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1OTM
 
 | | Calcium-binding mutant of the internalin B LRR domain | | Descriptor: | internalin B | | Authors: | Marino, M, Copp, J, Dramsi, S, Chapman, T, van der Geer, P, Cossart, P, Ghosh, P. | | Deposit date: | 2003-03-21 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Characterization of the calcium-binding sites of Listeria monocytogenes InlB
Biochem.Biophys.Res.Commun., 316, 2004
|
|
2MHZ
 
 | | Structure of Exocyclic S,S N6,N6-(2,3-Dihydroxy-1,4-butadiyl)-2'-Deoxyadenosine Adduct Induced by 1,2,3,4-Diepoxybutane in DNA | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(SDE)P*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3' | | Authors: | Kowal, E.A, Seneviratne, U, Wickramaratne, S, Doherty, K.E, Cao, X, Tretyakova, N, Stone, M.P. | | Deposit date: | 2013-12-05 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of Exocyclic R,R- and S,S-N(6),N(6)-(2,3-Dihydroxybutan-1,4-diyl)-2'-Deoxyadenosine Adducts Induced by 1,2,3,4-Diepoxybutane.
Chem.Res.Toxicol., 27, 2014
|
|
2MHX
 
 | | Structure of Exocyclic R,R N6,N6-(2,3-Dihydroxy-1,4-butadiyl)-2'-Deoxyadenosine Adduct Induced by 1,2,3,4-Diepoxybutane in DNA | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(RBD)P*AP*GP*AP*AP*G)-3'), 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3') | | Authors: | Kowal, E.A, Seneviratne, U, Wickramaratne, S, Doherty, K.E, Cao, X, Tretyakova, N, Stone, M.P. | | Deposit date: | 2013-12-05 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of Exocyclic R,R- and S,S-N(6),N(6)-(2,3-Dihydroxybutan-1,4-diyl)-2'-Deoxyadenosine Adducts Induced by 1,2,3,4-Diepoxybutane.
Chem.Res.Toxicol., 27, 2014
|
|
2PBE
 
 | |
2PBR
 
 | | Crystal structure of thymidylate kinase (aq_969) from Aquifex Aeolicus VF5 | | Descriptor: | SULFATE ION, Thymidylate kinase | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Nakagawa, N, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of thymidylate kinase (aq_969) from Aquifex Aeolicus VF5
To be Published
|
|
2PCN
 
 | | Crystal structure of S-adenosylmethionine: 2-dimethylmenaquinone methyltransferase (gk_1813) from geobacillus kaustophilus HTA426 | | Descriptor: | ACETATE ION, S-adenosylmethionine:2-demethylmenaquinone methyltransferase | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Rafi, Z.A, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of S-adenosylmethionine:2-dimethylmenaquinone methyltransferase (gk_1813) from geobacillus kaustophilus HTA426
To be Published
|
|
3DQY
 
 | | Crystal structure of Toluene 2,3-Dioxygenase Ferredoxin | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Toluene 1,2-dioxygenase system ferredoxin subunit | | Authors: | Friemann, R, Lee, K, Brown, E.N, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2008-07-10 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structures of the multicomponent Rieske non-heme iron toluene 2,3-dioxygenase enzyme system
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3DNL
 
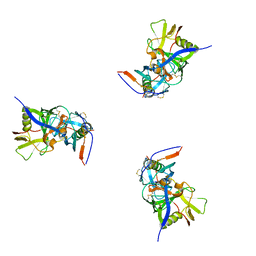 | | Molecular structure for the HIV-1 gp120 trimer in the b12-bound state | | Descriptor: | HIV-1 envelope glycoprotein gp120 | | Authors: | Borgnia, M.J, Liu, J, Bartesaghi, A, Sapiro, G, Subramaniam, S. | | Deposit date: | 2008-07-02 | | Release date: | 2008-08-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Molecular architecture of native HIV-1 gp120 trimers.
Nature, 455, 2008
|
|
1SQZ
 
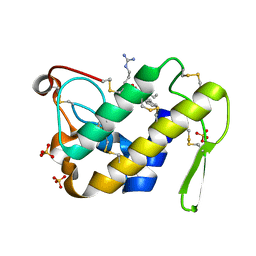 | | Design of specific inhibitors of Phopholipase A2: Crystal structure of the complex formed between Group II Phopholipase A2 and a designed peptide Dehydro-Ile-Ala-Arg-Ser at 1.2A resolution | | Descriptor: | Phospholipase A2, SULFATE ION, synthetic peptide | | Authors: | Singh, N, Prem Kumar, R, Somvanshi, R.K, Bilgrami, S, Ethayathulla, A.S, Sharma, S, Dey, S, Singh, T.P. | | Deposit date: | 2004-03-22 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Design of specific inhibitors of Phopholipase A2: Crystal structure of the complex formed between GroupII Phopholipase A2 and a designed peptide Dehydro-Ile-Ala-Arg-Ser at 1.2A resolution
To be Published
|
|
3JCF
 
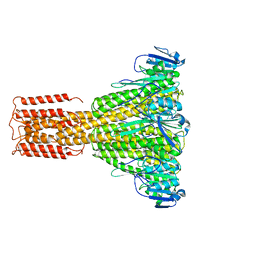 | |
3JQM
 
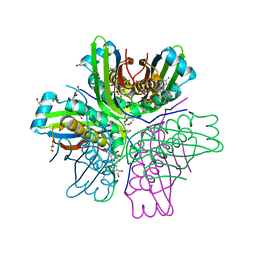 | | Binding of 5'-GTP to molybdenum cofactor biosynthesis protein MoaC from Thermus theromophilus HB8 | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Nakagawa, N, Sekar, K, Shinkai, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-07 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of apo and GTP-bound molybdenum cofactor biosynthesis protein MoaC from Thermus thermophilus HB8
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3JCH
 
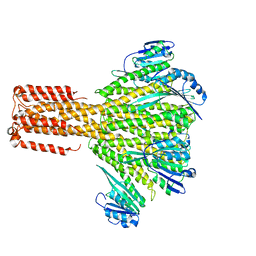 | |
3J7H
 
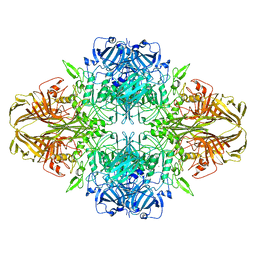 | | Structure of beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy | | Descriptor: | Beta-galactosidase, MAGNESIUM ION | | Authors: | Bartesaghi, A, Matthies, D, Banerjee, S, Merk, A, Subramaniam, S. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of beta-galactosidase at 3.2- angstrom resolution obtained by cryo-electron microscopy.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1SXK
 
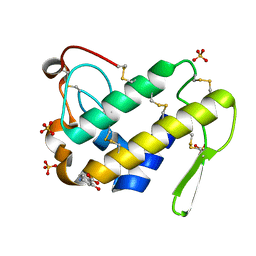 | | Crystal Structure of a complex formed between phospholipase A2 and a non-specific anti-inflammatory amino salicylic acid at 1.2 A resolution | | Descriptor: | 2-HYDROXY-4-AMINOBENZOIC ACID, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Singh, N, Bilgrami, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2004-03-31 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystal Structure of a complex formed between phospholipase A2 and a non-specific anti-inflammatory amino salicylic acid at 1.2 A resolution
To be Published
|
|
1T3C
 
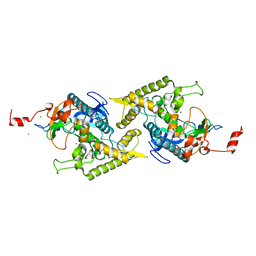 | | Clostridium botulinum type E catalytic domain E212Q mutant | | Descriptor: | CHLORIDE ION, ZINC ION, neurotoxin type E | | Authors: | Agarwal, R, Eswaramoorthy, S, Kumaran, D, Binz, T, Swaminathan, S. | | Deposit date: | 2004-04-26 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of botulinum neurotoxin type E catalytic domain and its mutant Glu212-->Gln reveals the pivotal role of the Glu212 carboxylate in the catalytic pathway
Biochemistry, 43, 2004
|
|
2P5U
 
 | | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase | | Authors: | Fu, Z.-Q, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhao, M, Dillard, B, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD
To be Published
|
|
2P9M
 
 | | Crystal structure of conserved hypothetical protein MJ0922 from Methanocaldococcus jannaschii DSM 2661 | | Descriptor: | Hypothetical protein MJ0922 | | Authors: | Zhao, M, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhu, J, Swindell II, J.T, Chen, L, Fu, Z.-Q, Charz, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of conserved hypothetical protein MJ0922 from Methanocaldococcus jannaschii DSM 2661
To be Published
|
|
2YOJ
 
 | | HCV NS5B polymerase complexed with pyridonylindole compound | | Descriptor: | 4-fluoranyl-6-[(7-fluoranyl-4-oxidanylidene-3H-quinazolin-6-yl)methyl]-8-(2-oxidanylidene-1H-pyridin-3-yl)furo[2,3-e]indole-7-carboxylic acid, PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Chen, K.X, Venkatraman, S, Anilkumar, G.N, Zeng, Q, Lesburg, C.A, Vibulbhan, B, Yang, W, Velazquez, F, Chan, T.-Y, Bennett, F, Sannigrahi, M, Jiang, Y, Duca, J.S, Pinto, P, Gavalas, S, Huang, Y, Wu, W, Selyutin, O, Agrawal, S, Feld, B, Huang, H.-C, Li, C, Cheng, K.-C, Shih, N.-Y, Kozlowski, J.A, Rosenblum, S.B, Njoroge, F.G. | | Deposit date: | 2012-10-24 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of Sch 900188: A Potent Hepatitis C Virus Ns5B Polymerase Inhibitor Prodrug as a Development Candidate
Acs Med.Chem.Lett., 5, 2014
|
|
2HXP
 
 | | Crystal Structure of the human phosphatase (DUSP9) | | Descriptor: | Dual specificity protein phosphatase 9, PHOSPHATE ION | | Authors: | Madegowda, M, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-03 | | Release date: | 2006-08-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2PBP
 
 | | Crystal structure of ENOYL-CoA hydrates subunit I (gk_2039) from geobacillus kaustophilus HTA426 | | Descriptor: | Enoyl-CoA hydratase subunit I | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki, R.C, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of ENOYL-CoA hydrates subunit I (gk_2039) from geobacillus kaustophilus HTA426
To be Published
|
|
1QFJ
 
 | | CRYSTAL STRUCTURE OF NAD(P)H:FLAVIN OXIDOREDUCTASE FROM ESCHERICHIA COLI | | Descriptor: | GLYCEROL, PROTEIN (FLAVIN REDUCTASE) | | Authors: | Ingelman, M, Ramaswamy, S, Niviere, V, Fontecave, M, Eklund, H. | | Deposit date: | 1999-04-12 | | Release date: | 1999-06-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of NAD(P)H:flavin oxidoreductase from Escherichia coli.
Biochemistry, 38, 1999
|
|
2P3E
 
 | | Crystal structure of AQ1208 from Aquifex aeolicus | | Descriptor: | Diaminopimelate decarboxylase | | Authors: | Zhu, J, Swindell II, J.T, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Fu, Z.-Q, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-08 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | To be Published
To be Published
|
|
2P5Y
 
 | | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase | | Authors: | Fu, Z.-Q, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhu, J, Swindell, J.T, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD
To be Published
|
|
