1KJ1
 
 | | MANNOSE-SPECIFIC AGGLUTININ (LECTIN) FROM GARLIC (ALLIUM SATIVUM) BULBS COMPLEXED WITH ALPHA-D-MANNOSE | | Descriptor: | alpha-D-mannopyranose, lectin I, lectin II | | Authors: | Ramachandraiah, G, Chandra, N.R, Surolia, A, Vijayan, M. | | Deposit date: | 2001-12-04 | | Release date: | 2002-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Re-refinement using reprocessed data to improve the quality of the structure: a case study involving garlic lectin.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
8G62
 
 | | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004 | | Descriptor: | 3-methoxy-5-(1-methylpiperidin-4-yl)-N-[4-(pyrrolidine-1-sulfonyl)phenyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Luci, D, Kales, S, Simeonov, A, Rai, G, Drayman, N, Tay, S, Oakes, S, Rosner, M, Chen, B, Dulin, N, Solway, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004
To Be Published
|
|
7KNF
 
 | | 1.80A resolution structure of independent Phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Ce-1 NHOH) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, DTY-ASP-TYR-PRO-GLY-ASP-HIS-CYS-TYR-LEU-TYR-GLY-THR, SODIUM ION, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Weidmann, M, Dranchak, P, Aitha, M, Queme, B, Collmus, C.D, Kanter, L, Lamy, L, Tao, D, Rai, G, Suga, H, Inglese, J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-activity relationship of ipglycermide binding to phosphoglycerate mutases.
J.Biol.Chem., 296, 2021
|
|
7KNG
 
 | | 2.10A resolution structure of independent Phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Ce-2 Y7F) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, DTY-ASP-TYR-PRO-GLY-ASP-PHE-CYS-TYR-LEU-TYR-GLY-THR-CYS, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Weidmann, M, Dranchak, P, Aitha, M, Queme, B, Collmus, C.D, Kanter, L, Lamy, L, Tao, D, Rai, G, Suga, H, Inglese, J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity relationship of ipglycermide binding to phosphoglycerate mutases.
J.Biol.Chem., 296, 2021
|
|
3RVH
 
 | | Crystal Structure of JMJD2A Complexed with Inhibitor | | Descriptor: | 8-hydroxy-3-(piperazin-1-yl)quinoline-5-carboxylic acid, GLYCEROL, Lysine-specific demethylase 4A, ... | | Authors: | King, O.N.F, Maloney, D.J, Tumber, A, Rai, G, Jadhav, A, Clifton, I.J, Heightman, T.D, Simeonov, A, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Crystal Structure of JMJD2A Complexed with Inhibitor
To be Published
|
|
6QYL
 
 | | Structure of MBP-Mcl-1 in complex with compound 8a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZ8
 
 | | Structure of Mcl-1 in complex with compound 10d | | Descriptor: | (2~{R})-2-[5-(3-chloranyl-2-methyl-4-oxidanyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QYK
 
 | | Structure of MBP-Mcl-1 in complex with compound 7a | | Descriptor: | (2~{R})-2-[6-ethyl-5-(1~{H}-indol-5-yl)thieno[2,3-d]pyrimidin-4-yl]oxypropanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZ6
 
 | | Structure of Mcl-1 in complex with compound 8b | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QYN
 
 | | Structure of MBP-Mcl-1 in complex with compound 10d | | Descriptor: | (2~{R})-2-[5-(3-chloranyl-2-methyl-4-oxidanyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QYP
 
 | | Structure of Mcl-1 in complex with compound 13 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-5-(4-methylpiperazin-1-yl)-4-oxidanyl-phenyl]-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZ5
 
 | | Structure of Mcl-1 in complex with compound 8a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZ7
 
 | | Structure of MBP-Mcl-1 in complex with compound 8b | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZB
 
 | | Structure of Mcl-1 in complex with compound 8d | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(2-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QYO
 
 | | Structure of MBP-Mcl-1 in complex with compound 18a | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QXJ
 
 | | Structure of MBP-Mcl-1 in complex with compound 6a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-5-yl)thieno[2,3-d]pyrimidin-4-yl]amino]propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-07 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6GL8
 
 | | Crystal structure of Bcl-2 in complex with the novel orally active inhibitor S55746 | | Descriptor: | Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2, ~{N}-(4-hydroxyphenyl)-3-[6-[[(3~{S})-3-(morpholin-4-ylmethyl)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]-1,3-benzodioxol-5-yl]-~{N}-phenyl-5,6,7,8-tetrahydroindolizine-1-carboxamide | | Authors: | Casara, P, Davidson, J, Claperon, A, Le Toumelin-Braizat, G, Vogler, M, Bruno, A, Chanrion, M, Lysiak-Auvity, G, Le Diguarher, T, Starck, J.B, Chen, I, Whitehead, N, Graham, C, Matassova, N, Dokurno, P, Pedder, C, Wang, Y, Qiu, S, Girard, A.M, Schneider, E, Grave, F, Studeny, A, Guasconi, G, Rocchetti, F, Maiga, S, Henlin, J.M, Colland, F, Kraus-Berthier, L, Le Gouill, S, Dyer, M.J.S, Hubbard, R, Wood, M, Amiot, M, Cohen, G.M, Hickman, J.A, Morris, E, Murray, J, Geneste, O. | | Deposit date: | 2018-05-23 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | S55746 is a novel orally active BCL-2 selective and potent inhibitor that impairs hematological tumor growth.
Oncotarget, 9, 2018
|
|
4FBX
 
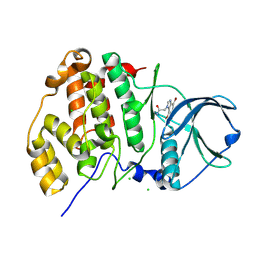 | | Complex structure of human protein kinase CK2 catalytic subunit crystallized in the presence of a bisubstrate inhibitor | | Descriptor: | CHLORIDE ION, Casein kinase II subunit alpha, bisubstrate inhibitor | | Authors: | Enkvist, E, Viht, K, Bischoff, N, Vahter, J, Saaver, S, Raidaru, G, Issinger, O.-G, Niefind, K, Uri, A. | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | A subnanomolar fluorescent probe for protein kinase CK2 interaction studies.
Org.Biomol.Chem., 10, 2012
|
|
5UMI
 
 | | Clostridium difficile TcdA-CROPs bound to PA50 Fab | | Descriptor: | PA50 Fab Heavy chain, PA50 Fab Light chain, Toxin A | | Authors: | Kroh, H.K, Chandrasekaran, R, Rosenthal, K, Woods, R, Jin, X, Ohi, M.D, Nyborg, A.C, Rainey, G.J, Warrener, P, Spiller, B.W, Lacy, D.B. | | Deposit date: | 2017-01-27 | | Release date: | 2017-07-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Use of a neutralizing antibody helps identify structural features critical for binding of Clostridium difficile toxin TcdA to the host cell surface.
J. Biol. Chem., 292, 2017
|
|
3H1Z
 
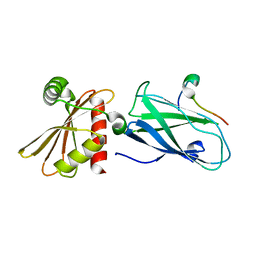 | | Molecular basis for the association of PIPKIgamma -p90 with the clathrin adaptor AP-2 | | Descriptor: | AP-2 complex subunit beta-1, Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma | | Authors: | Vahedi-Faridi, A, Kahlfeldt, N, Schaefer, J.G, Krainer, G, Keller, S, Saenger, W, Krauss, M, Haucke, V. | | Deposit date: | 2009-04-14 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Molecular basis for association of PIPKI gamma-p90 with clathrin adaptor AP-2.
J.Biol.Chem., 285, 2010
|
|
2VXY
 
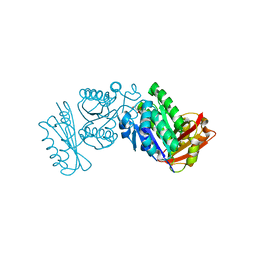 | | The structure of FTsZ from Bacillus subtilis at 1.7A resolution | | Descriptor: | CELL DIVISION PROTEIN FTSZ, CITRIC ACID, POTASSIUM ION | | Authors: | Barynin, V.V, Baker, P.J, Rice, D.W, Sedelnikova, S.E, Haydon, D.J, Stokes, N.R, Ure, R, Galbraith, G, Bennett, J.M, Brown, D.R, Heal, J.R, Sheridan, J.M, Aiwale, S.T, Chauhan, P.K, Srivastava, A, Taneja, A, Collins, I, Errington, J, Czaplewski, L.G. | | Deposit date: | 2008-07-15 | | Release date: | 2008-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An Inhibitor of Ftsz with Potent and Selective Anti-Staphylococcal Activity.
Science, 321, 2008
|
|
1DP3
 
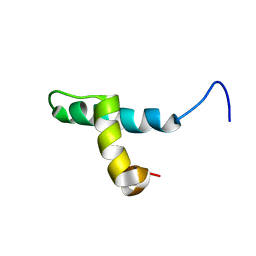 | | SOLUTION STRUCTURE OF THE DNA BINDING DOMAIN OF THE TRAM PROTEIN | | Descriptor: | TRAM PROTEIN | | Authors: | Stockner, T, Plugariu, C, Koraimann, G, Hoegenauer, G, Bermel, W, Prytulla, S, Sterk, H. | | Deposit date: | 1999-12-23 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of TraM.
Biochemistry, 40, 2001
|
|
1BWU
 
 | | MANNOSE-SPECIFIC AGGLUTININ (LECTIN) FROM GARLIC (ALLIUM SATIVUM) BULBS COMPLEXED WITH ALPHA-D-MANNOSE | | Descriptor: | PROTEIN (AGGLUTININ), alpha-D-mannopyranose | | Authors: | Chandra, N.R, Ramachandraiah, G, Bachhawat, K, Dam, T.K, Surolia, A, Vijayan, M. | | Deposit date: | 1998-09-28 | | Release date: | 1999-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a dimeric mannose-specific agglutinin from garlic: quaternary association and carbohydrate specificity.
J.Mol.Biol., 285, 1999
|
|
