2ZYH
 
 | | mutant A. Fulgidus lipase S136A complexed with fatty acid fragment | | Descriptor: | CALCIUM ION, HEXADECANE, Lipase, ... | | Authors: | Chen, C.K, Ko, T.P, Guo, R.T, Wang, A.H. | | Deposit date: | 2009-01-22 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of the alkalohyperthermophilic Archaeoglobus fulgidus lipase contains a unique C-terminal domain essential for long-chain substrate binding.
J.Mol.Biol., 390, 2009
|
|
2ZYR
 
 | | A. Fulgidus lipase with fatty acid fragment and magnesium | | Descriptor: | Lipase, putative, MAGNESIUM ION, ... | | Authors: | Chen, C.K, Ko, T.P, Guo, R.T, Wang, A.H. | | Deposit date: | 2009-01-28 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of the alkalohyperthermophilic Archaeoglobus fulgidus lipase contains a unique C-terminal domain essential for long-chain substrate binding.
J.Mol.Biol., 390, 2009
|
|
6IR2
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM2 at 1.4 Angstron resolution | | Descriptor: | MCherry fluorescent protein, mCherry's nanobody LaM2 | | Authors: | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | Deposit date: | 2018-11-09 | | Release date: | 2019-11-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.393 Å) | | Cite: | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
6IR1
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM4 at 1.9 Angstron resolution | | Descriptor: | MCherry fluorescent protein, mCherry's nanobody LaM4 | | Authors: | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | Deposit date: | 2018-11-09 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
3KZ0
 
 | | MCL-1 complex with MCL-1-specific selected peptide | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, Mcl-1 specific peptide MB7, SULFATE ION, ... | | Authors: | Dutta, S, Fire, E, Grant, R.A, Sauer, R.T, Keating, A.E. | | Deposit date: | 2009-12-07 | | Release date: | 2010-05-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Determinants of BH3 binding specificity for Mcl-1 versus Bcl-xL.
J.Mol.Biol., 398, 2010
|
|
3DU8
 
 | | Crystal structure of GSK-3 beta in complex with NMS-869553A | | Descriptor: | (7S)-2-(2-aminopyrimidin-4-yl)-7-(2-fluoroethyl)-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one, Glycogen synthase kinase-3 beta | | Authors: | Bossi, R.T. | | Deposit date: | 2008-07-17 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | First Cdc7 kinase inhibitors: pyrrolopyridinones as potent and orally active antitumor agents. 2. Lead discovery.
J.Med.Chem., 52, 2009
|
|
2LAH
 
 | | Solution NMR Structure of Mitotic checkpoint serine/threonine-protein kinase BUB1 N-terminal domain from Homo sapiens, Northeast Structural Genomics Consortium Target HR5460A (Methods Development) | | Descriptor: | Mitotic checkpoint serine/threonine-protein kinase BUB1 | | Authors: | Liu, G, Xiao, R, Lee, H, Hamilton, K, Acton, T.B, Ciccosanti, C, Everett, J.K, Shastry, R.T, Huang, Y.J, Montelione, G.T, Northeast Structural Genomics Consortium, n, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target HR5460A
To be Published
|
|
2M8W
 
 | | Restrained CS-Rosetta Solution NMR Structure of Staphylococcus aureus protein SAV1430. Northeast Structural Genomics Target ZR18. Structure determination | | Descriptor: | Uncharacterized protein | | Authors: | Mao, B, Tejero, R.T, Aramini, J.M, Snyder, D.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PDBStat: a universal restraint converter and restraint analysis software package for protein NMR.
J.Biomol.Nmr, 56, 2013
|
|
2M8X
 
 | | Restrained CS-Rosetta Solution NMR structure of the CARDB domain of PF1109 from Pyrococcus furiosus. Northeast Structural Genomics Consortium target PfR193A | | Descriptor: | Uncharacterized protein | | Authors: | Mao, B, Tejero, R.T, Aramini, J.M, Snyder, D.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PDBStat: a universal restraint converter and restraint analysis software package for protein NMR.
J.Biomol.Nmr, 56, 2013
|
|
7W1N
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_KRP | | Descriptor: | 1,2-ETHANEDIOL, BICINE, Leaf-branch compost cutinase | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7W44
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_RIP | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Leaf-branch compost cutinase | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7W45
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_KIP | | Descriptor: | CALCIUM ION, Leaf-branch compost cutinase, SODIUM ION | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
4A4O
 
 | | CRYSTAL STRUCTURE OF POLO-LIKE KINASE 1 IN COMPLEX WITH A 2-(2-AMINO- PYRIMIDIN-4-YL)-1,5,6,7-TETRAHYDRO-PYRROLOPYRIDIN-4-ONE INHIBITOR | | Descriptor: | 1-METHYL-2-(2-{[5-(4-METHYLPIPERAZIN-1-YL)-2-(TRIFLUOROMETHOXY)PHENYL]AMINO}PYRIMIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, L(+)-TARTARIC ACID, SERINE/THREONINE-PROTEIN KINASE PLK1, ... | | Authors: | Bertrand, J.A, Bossi, R.T. | | Deposit date: | 2011-10-18 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 5-(2-Amino-Pyrimidin-4-Yl)-1H-Pyrrole and 2-(2-Amino-Pyrimidin-4-Yl)-1,5,6,7-Tetrahydro-Pyrrolo[3,2-C]Pyridin-4-One Derivatives as New Classes of Selective and Orally Available Polo-Like Kinase 1 Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4A4L
 
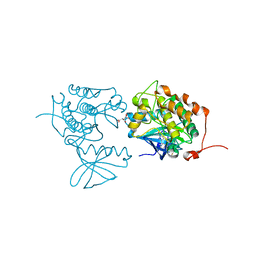 | | CRYSTAL STRUCTURE OF POLO-LIKE KINASE 1 IN COMPLEX WITH A 5-(2-AMINO- PYRIMIDIN-4-YL)-1H-PYRROLE INHIBITOR | | Descriptor: | 1-METHYL-5-(2-{[5-(4-METHYLPIPERAZIN-1-YL)-2-(TRIFLUOROMETHOXY)PHENYL]AMINO}PYRIMIDIN-4-YL)-1H-PYRROLE-3-CARBOXAMIDE, L(+)-TARTARIC ACID, SERINE/THREONINE-PROTEIN KINASE PLK1, ... | | Authors: | Bertrand, J.A, Bossi, R.T. | | Deposit date: | 2011-10-17 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 5-(2-Amino-Pyrimidin-4-Yl)-1H-Pyrrole and 2-(2-Amino-Pyrimidin-4-Yl)-1,5,6,7-Tetrahydro-Pyrrolo[3,2-C]Pyridin-4-One Derivatives as New Classes of Selective and Orally Available Polo-Like Kinase 1 Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7VVE
 
 | | Complex structure of a leaf-branch compost cutinase variant in complex with mono(2-hydroxyethyl) terephthalic acid | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 4-(2-hydroxyethyloxycarbonyl)benzoic acid, CALCIUM ION, ... | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-05 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7VVC
 
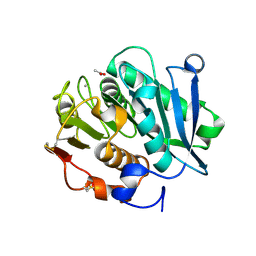 | | Crystal structure of inactive mutant of leaf-branch compost cutinase variant | | Descriptor: | ACETATE ION, ACETIC ACID, CALCIUM ION, ... | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-05 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
5XZO
 
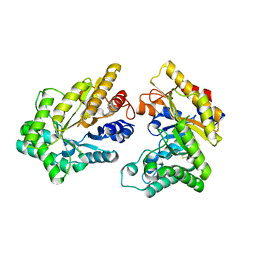 | | Crystal structure of GH10 xylanase XYL10C from Bispora. sp MEY-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-xylanase | | Authors: | You, S, Chen, C, Tu, T, Guo, R.T, Luo, H, Yao, B. | | Deposit date: | 2017-07-13 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of GH10 xylanase XYL10C from Bispora. sp MEY-1
To Be Published
|
|
3SD3
 
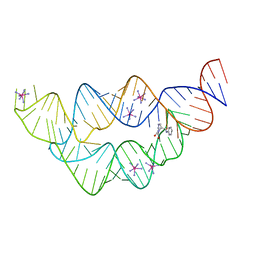 | | The structure of the tetrahydrofolate riboswitch containing a U25C mutation | | Descriptor: | IRIDIUM HEXAMMINE ION, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, Tetrahydrofolate riboswitch | | Authors: | Reyes, F.E, Trausch, J.J, Ceres, P, Batey, R.T. | | Deposit date: | 2011-06-08 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand binding sites in a single aptamer.
Structure, 19, 2011
|
|
3W02
 
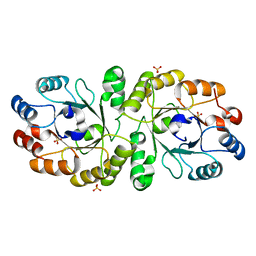 | | Crystal structure of PcrB complexed with SO4 from Staphylococcus aureus subsp. aureus Mu3 | | Descriptor: | Heptaprenylglyceryl phosphate synthase, SULFATE ION | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3W01
 
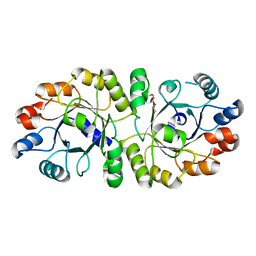 | | Crystal structure of PcrB complexed with PEG from Staphylococcus aureus subsp. aureus Mu3 | | Descriptor: | Heptaprenylglyceryl phosphate synthase, TRIETHYLENE GLYCOL | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3TSR
 
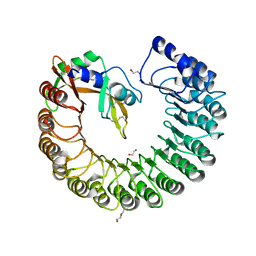 | | X-ray structure of mouse ribonuclease inhibitor complexed with mouse ribonuclease 1 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ribonuclease inhibitor, ... | | Authors: | Chang, A, Lomax, J.E, Bingman, C.A, Raines, R.T, Phillips Jr, G.N. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1999 Å) | | Cite: | Functional evolution of ribonuclease inhibitor: insights from birds and reptiles.
J.Mol.Biol., 426, 2014
|
|
3VNJ
 
 | | Crystal structures of D-Psicose 3-epimerase with D-psicose from Clostridium cellulolyticum H10 | | Descriptor: | D-psicose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | Authors: | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | Deposit date: | 2012-01-16 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3VZZ
 
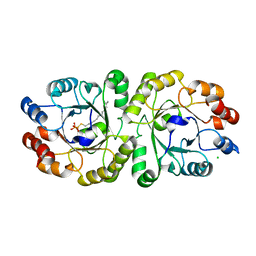 | | Crystal structure of PcrB complexed with FsPP from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION, ... | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VSV
 
 | | The complex structure of XylC with xylose | | Descriptor: | Xylosidase, alpha-D-xylopyranose, beta-D-xylopyranose | | Authors: | Huang, C.H, Sun, Y, Ko, T.P, Ma, Y, Chen, C.C, Zheng, Y, Chan, H.C, Pang, X, Wiegel, J, Shao, W, Guo, R.T. | | Deposit date: | 2012-05-09 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The substrate/product-binding modes of a novel GH120 beta-xylosidase (XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Biochem.J., 448, 2012
|
|
3VMV
 
 | | Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 | | Descriptor: | Pectate lyase, SULFATE ION | | Authors: | Zheng, Y, Huang, C.H, Liu, W, Ko, T.P, Xue, Y, Zhou, C, Zhang, G, Guo, R.T, Ma, Y. | | Deposit date: | 2011-12-16 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure and substrate-binding mode of a novel pectate lyase from alkaliphilic Bacillus sp. N16-5.
Biochem.Biophys.Res.Commun., 420, 2012
|
|
