5E5R
 
 | |
5DUO
 
 | | Crystal structure of native translocator protein 18kDa (TSPO) from Rhodobacter sphaeroides (A139T Mutant) in C2 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, FORMIC ACID, PROTOPORPHYRIN IX, ... | | Authors: | Li, F, Liu, J, Zheng, Y, Garavito, R.M, Ferguson-Miller, S. | | Deposit date: | 2015-09-20 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Response to Comment on "Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism".
Science, 350, 2015
|
|
5BNA
 
 | | THE PRIMARY MODE OF BINDING OF CISPLATIN TO A B-DNA DODECAMER: C-G-C-G-A-A-T-T-C-G-C-G | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PLATINUM TRIAMINE ION | | Authors: | Wing, R.M, Pjura, P, Drew, H.R, Dickerson, R.E. | | Deposit date: | 1983-08-22 | | Release date: | 1983-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The primary mode of binding of cisplatin to a B-DNA dodecamer: C-G-C-G-A-A-T-T-C-G-C-G
EMBO J., 3, 1984
|
|
5E01
 
 | | Crystal structure of HiNmlR, a MerR family regulator lacking the sensor domain, bound to palyndromic promoter DNA | | Descriptor: | 5'-D(*CP*TP*TP*AP*GP*AP*GP*TP*GP*CP*AP*CP*TP*CP*TP*AP*AP*G)-3', Uncharacterized HTH-type transcriptional regulator HI_0186 | | Authors: | Counago, R.M, Chang, C.W, Chen, N.H, Djoko, K.Y, McEwan, A.G, Kobe, B. | | Deposit date: | 2015-09-26 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of thiol-based regulation of formaldehyde detoxification in H. influenzae by a MerR regulator with no sensor region.
Nucleic Acids Res., 44, 2016
|
|
5DYE
 
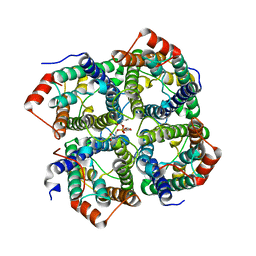 | | CRYSTAL STRUCTURE OF THE FULL LENGTH S156E MUTANT OF HUMAN AQUAPORIN 5 | | Descriptor: | Aquaporin-5, O-[(S)-{[(2S)-2-(hexanoyloxy)-3-(tetradecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-D-serine | | Authors: | Kitchen, P, Oeberg, F, Sjoehamn, J, Hedfalk, K, Bill, R.M, Conner, A.C, Conner, M.T, Toernroth-Horsefield, S. | | Deposit date: | 2015-09-24 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Plasma Membrane Abundance of Human Aquaporin 5 Is Dynamically Regulated by Multiple Pathways.
Plos One, 10, 2015
|
|
5E4Q
 
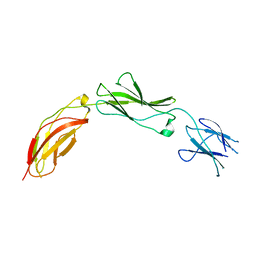 | |
4CSE
 
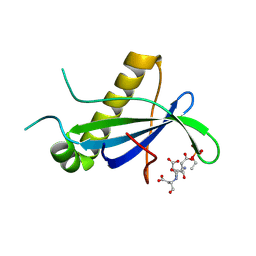 | | PIH N-terminal domain | | Descriptor: | PIH1 DOMAIN-CONTAINING PROTEIN 1, TELOMERE LENGTH REGULATION PROTEIN TEL2 HOMOLOG | | Authors: | Morgan, R.M, Roe, S.M. | | Deposit date: | 2014-03-07 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis for Phosphorylation-Dependent Recruitment of Tel2 to Hsp90 by Pih1.
Structure, 22, 2014
|
|
6EO6
 
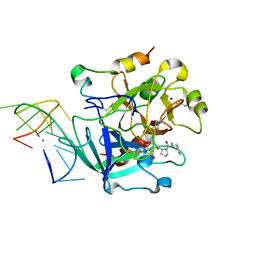 | | X-ray structure of the complex between human alpha-thrombin and modified 15-mer DNA aptamer containing 5-(3-(2-(1H-indol-3-yl)acetamide-N-yl)-1-propen-1-yl)-2'-deoxyuridine residue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GA63A - TBA MODIFIED APTAMER, ... | | Authors: | Dolot, R.M, Nawrot, B, Yang, X. | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity.
Nucleic Acids Res., 46, 2018
|
|
6S0B
 
 | | Crystal Structure of Properdin in complex with the CTC domain of C3/C3b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, Properdin, ... | | Authors: | van den Bos, R.M, Pearce, N.M, Gros, P. | | Deposit date: | 2019-06-14 | | Release date: | 2019-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Insights Into Enhanced Complement Activation by Structures of Properdin and Its Complex With the C-Terminal Domain of C3b.
Front Immunol, 10, 2019
|
|
6SF2
 
 | | Ternary complex of human bone morphogenetic protein 9 (BMP9) growth factor domain, its prodomain and extracellular domain of activin receptor-like kinase 1 (ALK1). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Growth/differentiation factor 2, Serine/threonine-protein kinase receptor R3 | | Authors: | Salmon, R.M, Guo, J, Yu, M, Li, W. | | Deposit date: | 2019-07-31 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular basis of ALK1-mediated signalling by BMP9/BMP10 and their prodomain-bound forms.
Nat Commun, 11, 2020
|
|
6SDV
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Formate reduced form | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, Formate dehydrogenase, ... | | Authors: | Oliveira, A.R, Mota, C, Mourato, C, Domingos, R.M, Santos, M.F.A, Gesto, D, Guigliarelli, B, Santos-Silva, T, Romao, M.J, Pereira, I.C. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards the mechanistic understanding of enzymatic CO2 reduction
Acs Catalysis, 2020
|
|
6SAL
 
 | | ROR(gamma)t ligand binding domain in complex with allosteric ligand FM26 | | Descriptor: | 4-[(~{E})-[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrrol-3-yl)-1,2-oxazol-4-yl]methylideneamino]benzoic acid, Nuclear receptor ROR-gamma | | Authors: | de Vries, R.M.J.M, Meijer, F.A, Doveston, R.G, Brunsveld, L. | | Deposit date: | 2019-07-17 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Ligand-Based Design of Allosteric Retinoic Acid Receptor-Related Orphan Receptor gamma t (ROR gamma t) Inverse Agonists.
J.Med.Chem., 63, 2020
|
|
4ECC
 
 | | Chimeric GST Containing Inserts of Kininogen Peptides | | Descriptor: | chimeric protein between GSHKT10 and domain 5 of kininogen-1 | | Authors: | Amber, A.B, Sergei, M.M, Yi, P, Rita, R, Xiaoping, Q, Marianne, P.-C, William, C.M, Vivien, Y, Keith, R.M, Anton, A.K. | | Deposit date: | 2012-03-26 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chimeric glutathione S-transferases containing inserts of kininogen peptides: potential novel protein therapeutics.
J.Biol.Chem., 287, 2012
|
|
6SF1
 
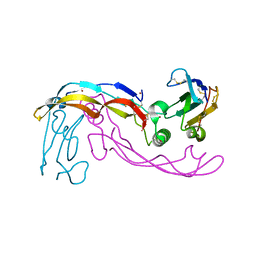 | | Bone morphogenetic protein 10 (BMP10) complexed with extracellular domain of activin receptor-like kinase 1 (ALK1). | | Descriptor: | Bone morphogenetic protein 10, NICKEL (II) ION, Serine/threonine-protein kinase receptor R3 | | Authors: | Salmon, R.M, Guo, J, Yu, M, Li, W. | | Deposit date: | 2019-07-30 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of ALK1-mediated signalling by BMP9/BMP10 and their prodomain-bound forms.
Nat Commun, 11, 2020
|
|
6SDR
 
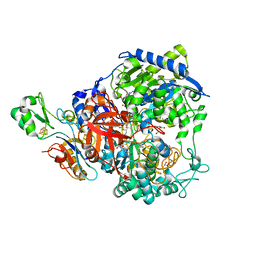 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Oxidized form | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Formate dehydrogenase, alpha subunit, ... | | Authors: | Oliveira, A.R, Mota, C, Mourato, C, Domingos, R.M, Santos, M.F.A, Gesto, D, Guigliarelli, B, Santos-Silva, T, Romao, M.J, Pereira, I.C. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Towards the mechanistic understanding of enzymatic CO2 reduction
Acs Catalysis, 2020
|
|
6S8F
 
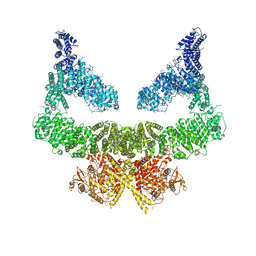 | | Structure of nucleotide-bound Tel1/ATM | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1 | | Authors: | Yates, L.A, Williams, R.M, Ayala, R, Zhang, X. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM Structure of Nucleotide-Bound Tel1ATMUnravels the Molecular Basis of Inhibition and Structural Rationale for Disease-Associated Mutations.
Structure, 28, 2020
|
|
6EBB
 
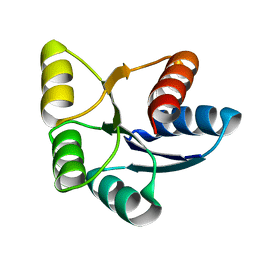 | |
6STU
 
 | | Adenovirus 30 Fiber Knob protein | | Descriptor: | 1,2-ETHANEDIOL, Fiber, GLUTAMIC ACID | | Authors: | Baker, A.T, Mundy, R.M, Rizkallah, P.J, Parker, A.L. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Broad sialic acid usage amongst species D human adenovirus
npj Viruses, 1, 2023
|
|
3O1C
 
 | | High resolution crystal structure of histidine triad nucleotide-binding protein 1 (Hint1) C38A mutant from rabbit complexed with Adenosine | | Descriptor: | ADENOSINE, Histidine triad nucleotide-binding protein 1, SODIUM ION | | Authors: | Dolot, R.M, Ozga, M, Krakowiak, A.K, Nawrot, B, Stec, W.J. | | Deposit date: | 2010-07-21 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Histidine Triad Nucleotide-binding Protein 1 (HINT-1) Phosphoramidase Transforms Nucleoside 5'-O-Phosphorothioates to Nucleoside 5'-O-Phosphates.
J.Biol.Chem., 285, 2010
|
|
3O1X
 
 | | High resolution crystal structure of histidine triad nucleotide-binding protein 1 (Hint1) C84A mutant from rabbit complexed with adenosine | | Descriptor: | ADENOSINE, Histidine triad nucleotide-binding protein 1, SODIUM ION | | Authors: | Dolot, R.M, Ozga, M, Krakowiak, A.K, Nawrot, B, Stec, W.J. | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Histidine Triad Nucleotide-binding Protein 1 (HINT-1) Phosphoramidase Transforms Nucleoside 5'-O-Phosphorothioates to Nucleoside 5'-O-Phosphates.
J.Biol.Chem., 285, 2010
|
|
6SXN
 
 | | Crystal structure of P212121 apo form of CrtE | | Descriptor: | Geranylgeranyl pyrophosphate synthase | | Authors: | Feng, Y, Morgan, R.M.L, Nixon, P.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase (CrtE) Involved in Cyanobacterial Terpenoid Biosynthesis.
Front Plant Sci, 11, 2020
|
|
3O1Z
 
 | | High resolution crystal structure of histidine triad nucleotide-binding protein 1 (Hint1) double cysteine mutant from rabbit | | Descriptor: | Histidine triad nucleotide-binding protein 1 | | Authors: | Dolot, R.M, Ozga, M, Krakowiak, A.K, Nawrot, B, Stec, W.J. | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Histidine Triad Nucleotide-binding Protein 1 (HINT-1) Phosphoramidase Transforms Nucleoside 5'-O-Phosphorothioates to Nucleoside 5'-O-Phosphates.
J.Biol.Chem., 285, 2010
|
|
6T13
 
 | | CRYSTAL STRUCTURE OF GLUCOCEREBROSIDASE IN COMPLEX WITH A PYRROLOPYRAZINE | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[2-(4-methoxyphenyl)-5-methyl-pyrrolo[2,3-b]pyrazin-6-yl]piperidin-1-yl]ethanone, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Benz, J, Ehler, A, Hug, M, Huber, S, Rufer, A.C, Guba, W, Jagasia, R, Hofmann, E.C, Rodriguez Sarmiento, R.M. | | Deposit date: | 2019-10-03 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Novel beta-Glucocerebrosidase Activators That Bind to a New Pocket at a Dimer Interface and Induce Dimerization.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
1PFF
 
 | | Crystal Structure of Homocysteine alpha-, gamma-lyase at 1.8 Angstroms | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, methionine gamma-lyase | | Authors: | Allen, T.W, Sridhar, V, Prasad, S.G, Han, Q, Xu, M, Tan, Y, Hoffman, R.M, Ramaswamy, S. | | Deposit date: | 2003-05-26 | | Release date: | 2004-08-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: |
|
|
6SZD
 
 | | Hydrogenase-2 variant R479K - hydrogen reduced form | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Beaton, S.E, Evans, R.M, Armstrong, F.A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hydrogen activation by NiFe-hydrogenases - consolidating the role of the pendant arginine.
To Be Published
|
|
