4MPN
 
 | | Crystal structure of pyruvate dehydrogenase kinase isoform 2 in complex with inhibitor PS10 | | Descriptor: | 2-[(2,4-dihydroxyphenyl)sulfonyl]-2,3-dihydro-1H-isoindole-4,6-diol, L(+)-TARTARIC ACID, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 2, ... | | Authors: | Gui, W.J, Tso, S.C, Chuang, J.L, Wu, C.Y, Qi, X, Tambar, U.K, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2013-09-13 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Structure-guided Development of Specific Pyruvate Dehydrogenase Kinase Inhibitors Targeting the ATP-binding Pocket.
J.Biol.Chem., 289, 2014
|
|
1MUO
 
 | | CRYSTAL STRUCTURE OF AURORA-2, AN ONCOGENIC SERINE-THREONINE KINASE | | Descriptor: | ADENOSINE, Aurora-related kinase 1 | | Authors: | Cheetham, G.M.T, Knegtel, R.M.A, Coll, J.T, Renwick, S.B, Swenson, L, Weber, P, Lippke, J.A, Austen, D.A. | | Deposit date: | 2002-09-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Aurora-2, an Oncogenic Serine/Threonine Kinase
J.Biol.Chem., 277, 2002
|
|
1N18
 
 | | Thermostable mutant of Human Superoxide Dismutase, C6A, C111S | | Descriptor: | COPPER (I) ION, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Cardoso, R.M.F, Thayer, M.M, DiDonato, M, Lo, T.P, Bruns, C.K, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2002-10-16 | | Release date: | 2002-11-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into Lou Gehrig's disease from the structure and instability of the A4V mutant of human Cu,Zn superoxide dismutase.
J.Mol.Biol., 324, 2002
|
|
4V5Q
 
 | | The crystal structure of EF-Tu and G24A-tRNA-Trp bound to a near- cognate codon on the 70S ribosome | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Schmeing, T.M, Voorhees, R.M, Ramakrishnan, V. | | Deposit date: | 2010-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How Mutations in tRNA Distant from the Anticodon Affect the Fidelity of Decoding.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4V5E
 
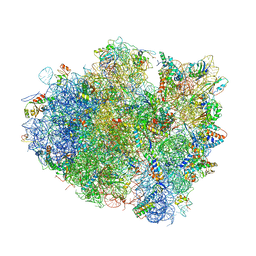 | | Insights into translational termination from the structure of RF2 bound to the ribosome | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Weixlbaumer, A, Jin, H, Neubauer, C, Voorhees, R.M, Petry, S, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-04-30 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Insights Into Translational Termination from the Structure of Rf2 Bound to the Ribosome.
Science, 322, 2008
|
|
1MZJ
 
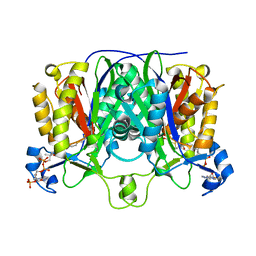 | | Crystal Structure of the Priming beta-Ketosynthase from the R1128 Polyketide Biosynthetic Pathway | | Descriptor: | ACETYL GROUP, Beta-ketoacylsynthase III, COENZYME A | | Authors: | Pan, H, Tsai, S.C, Meadows, E.S, Miercke, L.J.W, Keatinge-Clay, A, O'Connell, J, Khosla, C, Stroud, R.M. | | Deposit date: | 2002-10-08 | | Release date: | 2002-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the priming beta-ketosynthase from the
R1128 polyketide biosynthetic pathway
Structure, 10
|
|
4LAB
 
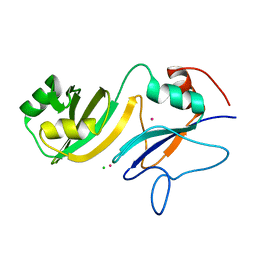 | | Crystal structure of the catalytic domain of RluB | | Descriptor: | CHLORIDE ION, PLATINUM (II) ION, Ribosomal large subunit pseudouridine synthase B | | Authors: | Czudnochowski, N, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2013-06-19 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5043 Å) | | Cite: | The mechanism of pseudouridine synthases from a covalent complex with RNA, and alternate specificity for U2605 versus U2604 between close homologs.
Nucleic Acids Res., 42, 2014
|
|
4W4T
 
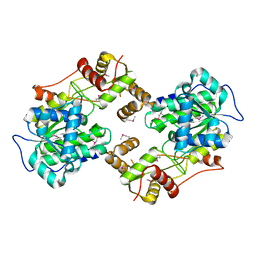 | | The crystal structure of the terminal R domain from the myxalamid PKS-NRPS biosynthetic pathway | | Descriptor: | ACETATE ION, MxaA | | Authors: | Tsai, S.C, Keasling, J.D, Luo, R, Barajas, J.F, Phelan, R.M, Schaub, A.J, Kliewer, J. | | Deposit date: | 2014-08-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Comprehensive Structural and Biochemical Analysis of the Terminal Myxalamid Reductase Domain for the Engineered Production of Primary Alcohols.
Chem.Biol., 22, 2015
|
|
1N1Z
 
 | | (+)-Bornyl Diphosphate Synthase: Complex with Mg and pyrophosphate | | Descriptor: | (+)-bornyl diphosphate synthase, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Whittington, D.A, Wise, M.L, Urbansky, M, Coates, R.M, Croteau, R.B, Christianson, D.W. | | Deposit date: | 2002-10-21 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bornyl diphosphate synthase: Structure and strategy for carbocation manipulation by a terpenoid cyclase
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4V5D
 
 | | Structure of the Thermus thermophilus 70S ribosome in complex with mRNA, paromomycin, acylated A- and P-site tRNAs, and E-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Weixlbaumer, A, Loakes, D, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome.
Nat. Struct. Mol. Biol., 16, 2009
|
|
4V5C
 
 | | Structure of the Thermus thermophilus 70S ribosome in complex with mRNA, paromomycin, acylated A-site tRNA, deacylated P-site tRNA, and E-site tRNA. | | Descriptor: | 16S ribosomal RNA, 23S Ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Weixlbaumer, A, Loakes, D, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Insights Into Substrate Stabilization from Snapshots of the Peptidyl Transferase Center of the Intact 70S Ribosome
Nat.Struct.Mol.Biol., 16, 2009
|
|
4V5R
 
 | | The crystal structure of EF-Tu and Trp-tRNA-Trp bound to a cognate codon on the 70S ribosome. | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Schmeing, T.M, Voorhees, R.M, Ramakrishnan, V. | | Deposit date: | 2010-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How Mutations in tRNA Distant from the Anticodon Affect the Fidelity of Decoding.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4LGT
 
 | |
4UTP
 
 | | Crystal structure of pneumococcal surface antigen PsaA in the Cd- bound, closed state | | Descriptor: | CADMIUM ION, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Luo, Z, Counago, R.M, Maher, M, Kobe, B. | | Deposit date: | 2014-07-22 | | Release date: | 2014-08-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae.
Nat Commun, 6, 2015
|
|
4V5S
 
 | | The crystal structure of EF-Tu and G24A-tRNA-Trp bound to a cognate codon on the 70S ribosome. | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Schmeing, T.M, Voorhees, R.M, Ramakrishnan, V. | | Deposit date: | 2010-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How Mutations in tRNA Distant from the Anticodon Affect the Fidelity of Decoding.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4UTO
 
 | | Crystal structure of pneumococcal surface antigen PsaA D280N in the Cd-bound, open state | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Luo, Z, Counago, R.M, Maher, M, Kobe, B. | | Deposit date: | 2014-07-22 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae.
Nat Commun, 6, 2015
|
|
4LK3
 
 | | Crystal structure of Human UDP-xylose synthase R236A substitution | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYROPHOSPHATE 2-, SULFATE ION, ... | | Authors: | Walsh Jr, R.M, Polizzi, S.J, Wood, Z.A. | | Deposit date: | 2013-07-05 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Man o' war mutation in UDP-alpha-D-xylose synthase favors the abortive catalytic cycle and uncovers a latent potential for hexamer formation.
Biochemistry, 54, 2015
|
|
4NJK
 
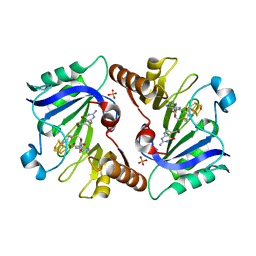 | | Crystal Structure of QueE from Burkholderia multivorans in complex with AdoMet, 7-carboxy-7-deazaguanine, and Mg2+ | | Descriptor: | 2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carboxylic acid, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Bruender, N.A, Young, A.P, McCarty, R.M, Bandarian, V, Drennan, C.L. | | Deposit date: | 2013-11-10 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism.
Nat.Chem.Biol., 10, 2014
|
|
7NSN
 
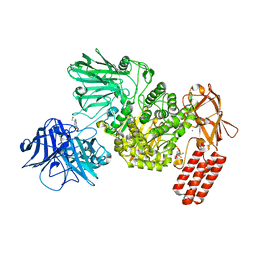 | | Multi-domain GH92 alpha-1,2-mannosidase from Neobacillus novalis: mannoimidazole complex | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Kolaczkowski, B.M, Moroz, O.V, Blagova, E, Davies, G.J, Wilson, K.S, Moeler, M.S, Meyer, A.S, Westh, P, Jensen, K, Krogh, K.B.R.M. | | Deposit date: | 2021-03-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural and functional characterization of a multi-domain GH92 alpha-1,2-mannosidase from Neobacillus novalis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7O7U
 
 | | Crystal structure of rsEGFP2 in the non-fluorescent off-state determined by serial femtosecond crystallography at room temperature | | Descriptor: | Green fluorescent protein | | Authors: | Hadjidemetriou, K, Woodhouse, J, Coquelle, N, Barends, T.R.M, Schlichting, I, Weik, M, Colletier, J.-P. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
7O7W
 
 | | Crystal structure of rsEGFP2 mutant V151L in the non-fluorescent off-state the determined by serial femtosecond crystallography at room temperature | | Descriptor: | Green fluorescent protein | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, Schlichting, I, Colletier, J.-P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
7O7V
 
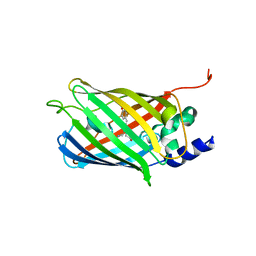 | | Crystal structure of rsEGFP2 mutant V151A in the fluorescent on-state determined by serial femtosecond crystallography at room temperature | | Descriptor: | Green fluorescent protein | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, Schlichting, I, Colletier, J.-P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
7O7X
 
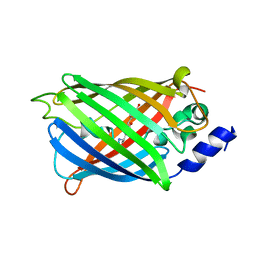 | | Crystal structure of rsEGFP2 mutant V151A in the non-fluorescent off-state determined by serial femtosecond crystallography at room temperature | | Descriptor: | Green fluorescent protein | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, Schlichting, I, Colletier, J.-P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
7OW0
 
 | |
7OXS
 
 | |
