2IHA
 
 | | Amidated variant of CMrVIA conotoxin | | Descriptor: | Lambda-conotoxin CMrVIA | | Authors: | Kini, R.M, Kang, T.S. | | Deposit date: | 2006-09-26 | | Release date: | 2007-08-14 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins
Biochemistry, 46, 2007
|
|
2II5
 
 | | Crystal structure of a cubic core of the dihydrolipoamide acyltransferase (E2b) component in the branched-chain alpha-ketoacid dehydrogenase complex (BCKDC), Isobutyryl-Coenzyme A-bound form | | Descriptor: | ACETATE ION, CHLORIDE ION, ISOBUTYRYL-COENZYME A, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Brautigam, C.A, Custorio, M, Chuang, D.T. | | Deposit date: | 2006-09-27 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain alpha-ketoacid dehydrogenase complex.
Embo J., 25, 2006
|
|
2IFI
 
 | | Ala6 Variant of ImI Conotoxin | | Descriptor: | Alpha-conotoxin ImI | | Authors: | Kini, R.M, Kang, T.S. | | Deposit date: | 2006-09-21 | | Release date: | 2007-08-14 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins
Biochemistry, 46, 2007
|
|
8DQT
 
 | | Human PDK1 kinase domain in complex with Valsartan | | Descriptor: | (2~{S})-3-methyl-2-[pentanoyl-[[4-[2-(2~{H}-1,2,3,4-tetrazol-5-yl)phenyl]phenyl]methyl]amino]butanoic acid, 3-phosphoinositide-dependent protein kinase 1, GLYCEROL | | Authors: | Gross, L.Z.F, Klinke, S, Biondi, R.M. | | Deposit date: | 2022-07-19 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Modulation of the substrate specificity of the kinase PDK1 by distinct conformations of the full-length protein.
Sci.Signal., 16, 2023
|
|
1YYS
 
 | | Y305F Trichodiene Synthase: Complex With Mg, Pyrophosphate, and (4S)-7-azabisabolene | | Descriptor: | (1S)-N,4-DIMETHYL-N-(4-METHYLPENT-3-ENYL)CYCLOHEX-3-ENAMINIUM, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Vedula, L.S, Rynkiewicz, M.J, Pyun, H.J, Coates, R.M, Cane, D.E, Christianson, D.W. | | Deposit date: | 2005-02-25 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular Recognition of the Substrate Diphosphate Group Governs Product Diversity in Trichodiene Synthase Mutants.
Biochemistry, 44, 2005
|
|
2IFZ
 
 | | Lys6 Variant of ImI Conotoxin | | Descriptor: | Alpha-conotoxin ImI | | Authors: | Kini, R.M, Kang, T.S. | | Deposit date: | 2006-09-22 | | Release date: | 2007-08-14 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins
Biochemistry, 46, 2007
|
|
1ZMO
 
 | | Apo structure of haloalcohol dehalogenase HheA of Arthrobacter sp. AD2 | | Descriptor: | halohydrin dehalogenase | | Authors: | de Jong, R.M, Kalk, K.H, Tang, L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2005-05-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray structure of the haloalcohol dehalogenase HheA from Arthrobacter sp. strain AD2: insight into enantioselectivity and halide binding in the haloalcohol dehalogenase family.
J.Bacteriol., 188, 2006
|
|
2IHW
 
 | | Crystal structure of a cubic core of the dihydrolipoamide acyltransferase (E2b) component in the branched-chain alpha-ketoacid dehydrogenase complex (BCKDC), apo form | | Descriptor: | ACETATE ION, CHLORIDE ION, Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Brautigam, C.A, Custorio, M, Chuang, D.T. | | Deposit date: | 2006-09-27 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain alpha-ketoacid dehydrogenase complex.
Embo J., 25, 2006
|
|
1Z1Q
 
 | |
2IS9
 
 | | Structure of yeast DCN-1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Defective in cullin neddylation protein 1, ... | | Authors: | Yang, X, Zhou, J, Sun, L, Wei, Z, Gao, J, Gong, W, Xu, R.M, Rao, Z, Liu, Y. | | Deposit date: | 2006-10-16 | | Release date: | 2007-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for the function of DCN-1 in protein Neddylation.
J.Biol.Chem., 282, 2007
|
|
1Y4C
 
 | | Designed Helical Protein fusion MBP | | Descriptor: | Maltose binding protein fused with designed helical protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | LaPorte, S.L, Forsyth, C.M, Cunningham, B.C, Miercke, L.J, Akhavan, D, Stroud, R.M. | | Deposit date: | 2004-11-30 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novo design of an IL-4 antagonist and its structure at 1.9 A.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1Y6B
 
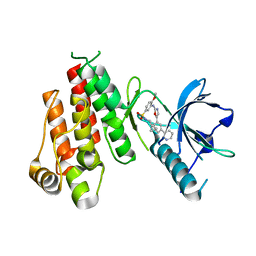 | | Crystal structure of VEGFR2 in complex with a 2-anilino-5-aryl-oxazole inhibitor | | Descriptor: | N-(CYCLOPROPYLMETHYL)-4-(METHYLOXY)-3-({5-[3-(3-PYRIDINYL)PHENYL]-1,3-OXAZOL-2-YL}AMINO)BENZENESULFONAMIDE, Vascular endothelial growth factor receptor 2 | | Authors: | Harris, P.A, Cheung, M, Hunter, R.N, Brown, M.L, Veal, J.M, Nolte, R.T, Wang, L, Liu, W, Crosby, R.M, Johnson, J.H, Epperly, A.H, Kumar, R, Luttrell, D.K, Stafford, J.A. | | Deposit date: | 2004-12-05 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and evaluation of 2-anilino-5-aryloxazoles as a novel class of VEGFR2 kinase inhibitors.
J.Med.Chem., 48, 2005
|
|
2IH7
 
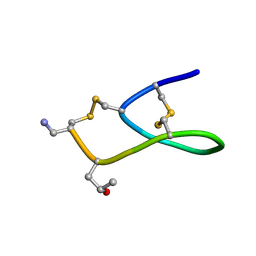 | | Amidated Pro6 Analogue of CMrVIA conotoxin | | Descriptor: | Lambda-conotoxin CMrVIA | | Authors: | Kini, R.M, Kang, T.S. | | Deposit date: | 2006-09-26 | | Release date: | 2007-08-14 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins
Biochemistry, 46, 2007
|
|
2MFN
 
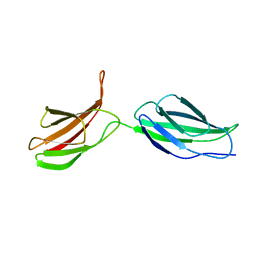 | | SOLUTION NMR STRUCTURE OF LINKED CELL ATTACHMENT MODULES OF MOUSE FIBRONECTIN CONTAINING THE RGD AND SYNERGY REGIONS, 10 STRUCTURES | | Descriptor: | FIBRONECTIN | | Authors: | Copie, V, Tomita, Y, Akiyama, S.K, Aota, S, Yamada, K.M, Venable, R.M, Pastor, R.W, Krueger, S, Torchia, D.A. | | Deposit date: | 1998-02-11 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of linked cell attachment modules of mouse fibronectin containing the RGD and synergy regions: comparison with the human fibronectin crystal structure.
J.Mol.Biol., 277, 1998
|
|
1Y8P
 
 | | Crystal structure of the PDK3-L2 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, ... | | Authors: | Kato, M, Chuang, J.L, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2004-12-13 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex.
Embo J., 24, 2005
|
|
1Y0S
 
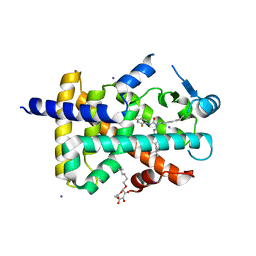 | | Crystal structure of PPAR delta complexed with GW2331 | | Descriptor: | (2S)-2-(4-[2-(3-[2,4-DIFLUOROPHENYL]-1-HEPTYLUREIDO)ETHYL]PHENOXY)-2-METHYLBUTYRIC ACID, IODIDE ION, Peroxisome proliferator activated receptor delta, ... | | Authors: | Takada, I, Yu, R.T, Xu, H.E, Xu, R.X, Lambert, M.H, Montana, V.G, Kliewer, S.A, Evans, R.M, Umesono, K. | | Deposit date: | 2004-11-16 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Alteration of a Single Amino Acid in Peroxisome Proliferator-Activated Receptor-alpha (PPARalpha) Generates a PPAR delta Phenotype
MOL.ENDOCRINOL., 14, 2000
|
|
6L3H
 
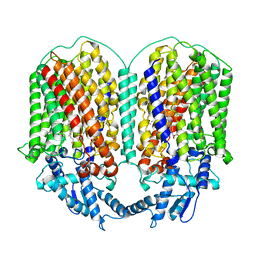 | | Cryo-EM structure of dimeric quinol dependent Nitric Oxide Reductase (qNOR) from the pathogen Neisseria meninigitidis | | Descriptor: | CALCIUM ION, FE (III) ION, Nitric-oxide reductase, ... | | Authors: | Jamali, M.M.A, Gopalasingam, C.C, Johnson, R.M, Tosha, T, Muench, S.P, Muramoto, K, Antonyuk, S.V, Shiro, Y, Hasnain, S.S. | | Deposit date: | 2019-10-11 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | The active form of quinol-dependent nitric oxide reductase fromNeisseria meningitidisis a dimer.
Iucrj, 7, 2020
|
|
4EGP
 
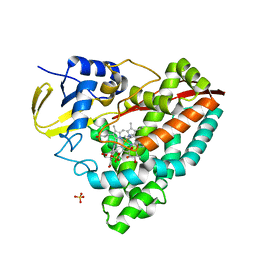 | | The X-ray crystal structure of CYP199A4 in complex with 2-naphthoic acid | | Descriptor: | CHLORIDE ION, Cytochrome P450, GLYCEROL, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Zhou, R.M, Tan, A.B.H, Wong, L.-L. | | Deposit date: | 2012-03-31 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Investigation of the substrate range of CYP199A4: modification of the partition between hydroxylation and desaturation activities by substrate and protein engineering
Chemistry, 18, 2012
|
|
2AQ9
 
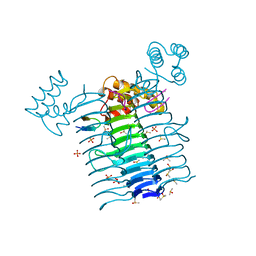 | | Structure of E. coli LpxA with a bound peptide that is competitive with acyl-ACP | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Williams, A.H, Immormino, R.M, Gewirth, D.T, Raetz, C.R. | | Deposit date: | 2005-08-17 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of UDP-N-acetylglucosamine acyltransferase with a bound antibacterial pentadecapeptide.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2B16
 
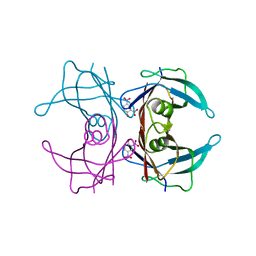 | | The crystal structure of 2,4-dinitrophenol in complex with the amyloidogenic variant Transthyretin Tyr78Phe | | Descriptor: | 2,4-DINITROPHENOL, Transthyretin | | Authors: | Morais-de-Sa, E, Neto-Silva, R.M, Pereira, P.J, Saraiva, M.J, Damas, A.M. | | Deposit date: | 2005-09-15 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The binding of 2,4-dinitrophenol to wild-type and amyloidogenic transthyretin
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2JJX
 
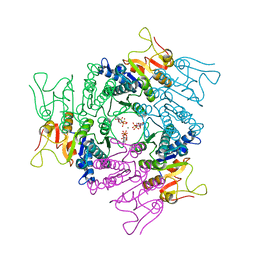 | | THE CRYSTAL STRUCTURE OF UMP KINASE FROM BACILLUS ANTHRACIS (BA1797) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, URIDYLATE KINASE | | Authors: | Meier, C, Carter, L.G, Mancini, E.J, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-04-23 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The Crystal Structure of Ump Kinase from Bacillus Anthracis (Ba1797) Reveals an Allosteric Nucleotide-Binding Site.
J.Mol.Biol., 381, 2008
|
|
6M9U
 
 | | Structure of the apo-form of 20beta-Hydroxysteroid Dehydrogenase from Bifidobacterium adolescentis strain L2-32 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Mythen, S.M, Pollet, R.M, Koropatkin, N.M, Ridlon, J.M. | | Deposit date: | 2018-08-24 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of 20 beta-hydroxysteroid dehydrogenase fromBifidobacterium adolescentisstrain L2-32.
J.Biol.Chem., 294, 2019
|
|
2JR3
 
 | | Antibacterial Peptide from Eggshell Matrix: Structure and Self-assembly of beta-defensin Like Peptide from the Chinese Soft-shelled Turtle Eggshell | | Descriptor: | Pelovaterin | | Authors: | Vivekanandan, S, Lakshminarayanan, R, Jois, S.D.S, Perumal Samy, R, Banerjee, Y, Chi-Jin, E.O, Teo, K.W, Kini, R.M, Valiyaveettil, S. | | Deposit date: | 2007-06-20 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure, self-assembly, and dual role of a beta-defensin-like peptide from the Chinese soft-shelled turtle eggshell matrix.
J.Am.Chem.Soc., 130, 2008
|
|
7N1W
 
 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
7N1Y
 
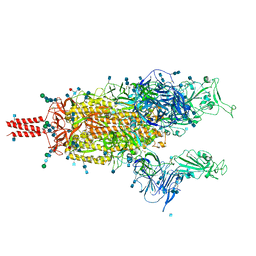 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
