1H9C
 
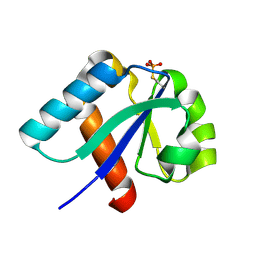 | | NMR structure of cysteinyl-phosphorylated enzyme IIB of the N,N'-diacetylchitobiose specific phosphoenolpyruvate-dependent phosphotransferase system of Escherichia coli. | | Descriptor: | PTS SYSTEM, CHITOBIOSE-SPECIFIC IIB COMPONENT | | Authors: | Ab, E, Schuurman-Wolters, G.K, Nijlant, D, Dijkstra, K, Saier, M.H, Robillard, G.T, Scheek, R.M. | | Deposit date: | 2001-03-07 | | Release date: | 2001-05-21 | | Last modified: | 2018-01-31 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Cysteinyl-Phosphorylated Enzyme Iib of the N,N'-Diacetylchitobiose Specific Phosphoenolpyruvate-Dependentphosphotransferase System of Escherichia Coli
J.Mol.Biol., 308, 2001
|
|
1HDN
 
 | |
6QU8
 
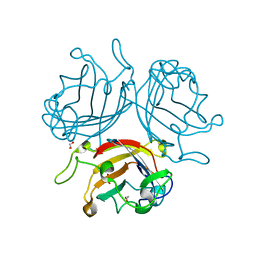 | | Adenovirus Serotype 26 (Ad26) in complex with sialic acid, pH8.0 | | Descriptor: | 1,2-ETHANEDIOL, Fiber, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Baker, A.T, Rizkallah, P.J, Parker, A.L, Mundy, R.M. | | Deposit date: | 2019-02-26 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Human adenovirus type 26 uses sialic acid-bearing glycans as a primary cell entry receptor.
Sci Adv, 5, 2019
|
|
6QQ5
 
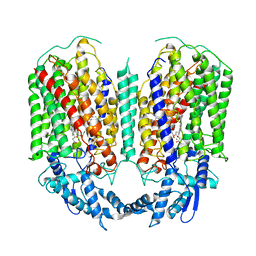 | | Cryo-EM structure of dimeric quinol dependent nitric oxide reductase (qNOR) from Alcaligenes xylosoxidans | | Descriptor: | CALCIUM ION, FE (III) ION, Nitric oxide reductase subunit B, ... | | Authors: | Gopalasingam, C.C, Johnson, R.M, Chiduza, G.N, Tosha, T, Yamamoto, M, Shiro, Y, Antonyuk, S.V, Muench, S.P, Hasnain, S.S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Dimeric structures of quinol-dependent nitric oxide reductases (qNORs) revealed by cryo-electron microscopy.
Sci Adv, 5, 2019
|
|
1IDE
 
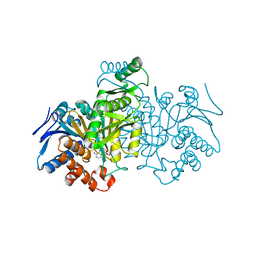 | | ISOCITRATE DEHYDROGENASE Y160F MUTANT STEADY-STATE INTERMEDIATE COMPLEX (LAUE DETERMINATION) | | Descriptor: | ISOCITRATE DEHYDROGENASE, ISOCITRIC ACID, MAGNESIUM ION, ... | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
6QU6
 
 | | Adenovirus Serotype 26 (Ad26) in complex with sialic acid, pH4.0 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Fiber, ... | | Authors: | Baker, A.T, Mundy, R.M, Parker, A.L, Rizkallah, P.J. | | Deposit date: | 2019-02-26 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Human adenovirus type 26 uses sialic acid-bearing glycans as a primary cell entry receptor.
Sci Adv, 5, 2019
|
|
6QQ6
 
 | | Cryo-EM structure of dimeric quinol dependent nitric oxide reductase (qNOR) Val495Ala mutant from Alcaligenes xylosoxidans | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Gopalasingam, C.C, Johnson, R.M, Chiduza, G.N, Tosha, T, Yamamoto, M, Shiro, Y, Antonyuk, S.V, Muench, S.P, Hasnain, S.S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dimeric structures of quinol-dependent nitric oxide reductases (qNORs) revealed by cryo-electron microscopy.
Sci Adv, 5, 2019
|
|
2J9F
 
 | | Human branched-chain alpha-ketoacid dehydrogenase-decarboxylase E1b | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXY-3-METHYL-PROPYL-THIAMIN DIPHOSPHATE, ... | | Authors: | Jun, L, Machius, M, Chuang, J.L, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2006-11-07 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Two Active Sites in Human Branched-Chain Alpha- Keto Acid Dehydrogenase Operate Independently without an Obligatory Alternating-Site Mechanism.
J.Biol.Chem., 282, 2007
|
|
2INY
 
 | |
2TGD
 
 | |
2TSC
 
 | | STRUCTURE, MULTIPLE SITE BINDING, AND SEGMENTAL ACCOMODATION IN THYMIDYLATE SYNTHASE ON BINDING D/UMP AND AN ANTI-FOLATE | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Montfort, W.R, Stroud, R.M. | | Deposit date: | 1991-07-03 | | Release date: | 1991-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure, multiple site binding, and segmental accommodation in thymidylate synthase on binding dUMP and an anti-folate.
Biochemistry, 29, 1990
|
|
1C0U
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH BM+50.0934 | | Descriptor: | (R)-(+) 5(9BH)-OXO-9B-PHENYL-2,3-DIHYDROTHIAZOLO[2,3-A]ISOINDOL-3-CARBOXYLIC ACID METHYL ESTER, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | Authors: | Ren, J, Esnouf, R.M, Hopkins, A.L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 1999-07-19 | | Release date: | 2000-07-19 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystallographic analysis of the binding modes of thiazoloisoindolinone non-nucleoside inhibitors to HIV-1 reverse transcriptase and comparison with modeling studies.
J.Med.Chem., 42, 1999
|
|
2BFE
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-12-06 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BFB
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-12-06 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BFD
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-12-06 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BFC
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-12-06 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BEU
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXY-3-METHYL-PROPYL-THIAMIN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-11-30 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BEW
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXYPHENYL-THIAMIN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-11-30 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BFF
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-12-06 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BEV
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXY-2-METHYL-BUTYL-THIAMIN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-11-30 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BBQ
 
 | | STRUCTURAL BASIS FOR RECOGNITION OF POLYGLUTAMYL FOLATES BY THYMIDYLATE SYNTHASE | | Descriptor: | 10-PARPARGYL-5,8-DIDEAZAFOLATE-4-GLUTAMIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Kamb, A, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1992-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition of polyglutamyl folates by thymidylate synthase.
Biochemistry, 31, 1992
|
|
1C0T
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH BM+21.1326 | | Descriptor: | (R)-(+)9B-(3-METHYL)PHENYL-2,3-DIHYDROTHIAZOLO[2,3-A]ISOINDOL-5(9BH)-ONE, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | Authors: | Ren, J, Esnouf, R.M, Hopkins, A.L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 1999-07-19 | | Release date: | 2000-07-19 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic analysis of the binding modes of thiazoloisoindolinone non-nucleoside inhibitors to HIV-1 reverse transcriptase and comparison with modeling studies.
J.Med.Chem., 42, 1999
|
|
1IDC
 
 | | ISOCITRATE DEHYDROGENASE FROM E.COLI (MUTANT K230M), STEADY-STATE INTERMEDIATE COMPLEX DETERMINED BY LAUE CRYSTALLOGRAPHY | | Descriptor: | 2-OXALOSUCCINIC ACID, ISOCITRATE DEHYDROGENASE, MAGNESIUM ION | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
1IDF
 
 | | ISOCITRATE DEHYDROGENASE K230M MUTANT APO ENZYME | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
3TMS
 
 | |
