2OQJ
 
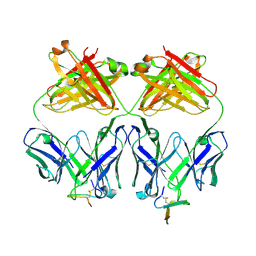 | | Crystal structure analysis of Fab 2G12 in complex with peptide 2G12.1 | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, peptide 2G12.1 (ACPPSHVLDMRSGTCLAAEGK) | | Authors: | Calarese, D.A, Stanfield, R.L, Menendez, A, Scott, J.K, Wilson, I.A. | | Deposit date: | 2007-01-31 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A peptide inhibitor of HIV-1 neutralizing antibody 2G12 is not a structural mimic of the natural carbohydrate epitope on gp120.
Faseb J., 22, 2008
|
|
7LEZ
 
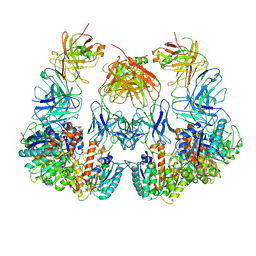 | | Trimeric human Arginase 1 in complex with mAb1 - 2 hArg:2 mAb1 complex | | Descriptor: | Arginase-1, MANGANESE (II) ION, mAb1 heavy chain, ... | | Authors: | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
7LEY
 
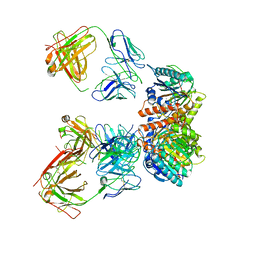 | | Trimeric human Arginase 1 in complex with mAb5 | | Descriptor: | Arginase-1, MANGANESE (II) ION, mAb5 heavy chain, ... | | Authors: | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
7LEX
 
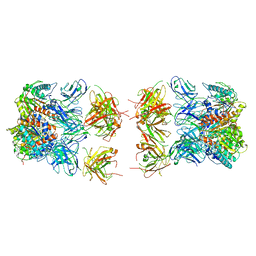 | |
7LF2
 
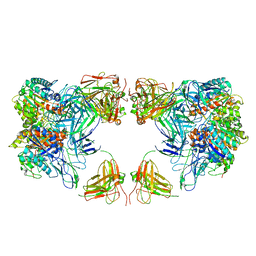 | | Trimeric human Arginase 1 in complex with mAb4 | | Descriptor: | Arginase-1, MANGANESE (II) ION, mAb4 monoclonal antibody heavy chain, ... | | Authors: | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
7LF0
 
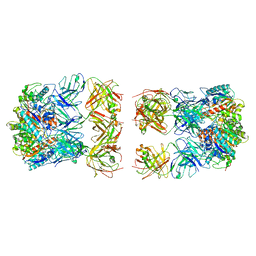 | |
7LF1
 
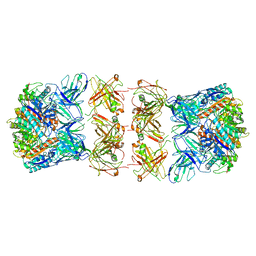 | | Trimeric human Arginase 1 in complex with mAb3 | | Descriptor: | Arginase-1, MANGANESE (II) ION, mAb3 heavy chain, ... | | Authors: | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
3VBN
 
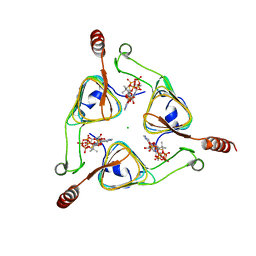 | |
6V7D
 
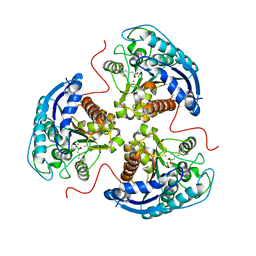 | | Human Arginase1 Complexed with Bicyclic Inhibitor Compound 10 | | Descriptor: | Arginase-1, MANGANESE (II) ION, {3-[(3aR,4R,5S,6aR)-4-azaniumyl-4-carboxyoctahydrocyclopenta[b]pyrrol-1-ium-5-yl]propyl}(trihydroxy)borate(1-) | | Authors: | Palte, R.L, Lesburg, C.A. | | Deposit date: | 2019-12-08 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery and Optimization of Rationally Designed Bicyclic Inhibitors of Human Arginase to Enhance Cancer Immunotherapy.
Acs Med.Chem.Lett., 11, 2020
|
|
6V7E
 
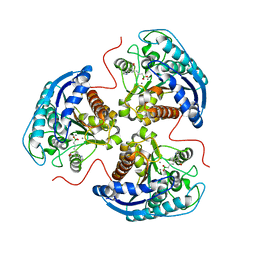 | | Human Arginase1 Complexed with Bicyclic Inhibitor Compound 12 | | Descriptor: | 3-[(5~{S},7~{S},8~{S})-8-azanyl-8-carboxy-1-azaspiro[4.4]nonan-7-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boranuide, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2019-12-08 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery and Optimization of Rationally Designed Bicyclic Inhibitors of Human Arginase to Enhance Cancer Immunotherapy.
Acs Med.Chem.Lett., 11, 2020
|
|
6VIO
 
 | |
2PLX
 
 | |
2OK0
 
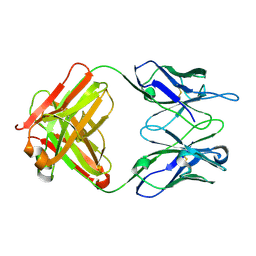 | | Fab ED10-DNA complex | | Descriptor: | 5'-D(*TP*C)-3', Fab ED10 heavy chain, Fab ED10 light chain | | Authors: | Stanfield, R.L, Sanguineti, S, Wilson, I.A, de Prat-Gay, G. | | Deposit date: | 2007-01-15 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Specific recognition of a DNA immunogen by its elicited antibody
J.Mol.Biol., 370, 2007
|
|
2OF1
 
 | |
2UZI
 
 | | Crystal structure of HRAS(G12V) - anti-RAS Fv complex | | Descriptor: | ANTI-RAS FV HEAVY CHAIN, ANTI-RAS FV LIGHT CHAIN, GTPASE HRAS, ... | | Authors: | Tanaka, T, williams, R.L, Rabbitts, T.H. | | Deposit date: | 2007-04-27 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tumour Prevention by a Single Antibody Domain Targeting the Interaction of Signal Transduction Proteins with Ras.
Embo J., 26, 2007
|
|
3UM0
 
 | |
2V6X
 
 | | Stractural insight into the interaction between ESCRT-III and Vps4 | | Descriptor: | DOA4-INDEPENDENT DEGRADATION PROTEIN 4, SULFATE ION, VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 4 | | Authors: | Obita, T, Perisic, O, Ghazi-Tabatabai, S, Saksena, S, Emr, S.D, Williams, R.L. | | Deposit date: | 2007-07-23 | | Release date: | 2007-10-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Basis for Selective Recognition of Escrt-III by the Aaa ATPase Vps4
Nature, 449, 2007
|
|
3VBK
 
 | | Crystal Structure of the S84A mutant of AntD, an N-acyltransferase from Bacillus cereus in complex with dTDP-4-amino-4,6-dideoxyglucose and Coenzyme A | | Descriptor: | BICARBONATE ION, CHLORIDE ION, COENZYME A, ... | | Authors: | Kubiak, R.L, Holden, H.M. | | Deposit date: | 2012-01-02 | | Release date: | 2012-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Studies of AntD: An N-Acyltransferase Involved in the Biosynthesis of d-Anthrose.
Biochemistry, 51, 2012
|
|
2V1Y
 
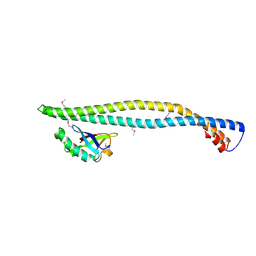 | | Structure of a phosphoinositide 3-kinase alpha adaptor-binding domain (ABD) in a complex with the iSH2 domain from p85 alpha | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY SUBUNIT ALPHA, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT ALPHA ISOFORM | | Authors: | Miled, N, Yan, Y, Hon, W.C, Perisic, O, Zvelebil, M, Inbar, Y, Schneidman-Duhovny, D, Wolfson, H.J, Backer, J.M, Williams, R.L. | | Deposit date: | 2007-05-30 | | Release date: | 2007-07-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of Two Classes of Cancer Mutations in the Phosphoinositide 3-Kinase Catalytic Subunit.
Science, 317, 2007
|
|
3TEO
 
 | | APO Form of carbon disulfide hydrolase (selenomethionine form) | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, Carbon disulfide hydrolase | | Authors: | Smeulders, M.J, Barends, T.R.M.B, Pol, A, Scherer, A, Zandvoort, M.H, Udvarhelyi, A, Khadem, A, Menzel, A, Hermans, J, Shoeman, R.L, Wessels, H.J.C.T, van den Heuvel, L.P, Russ, L, Schlichting, I, Jetten, M.S.M, Op den Camp, H.J.M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-10-19 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evolution of a new enzyme for carbon disulphide conversion by an acidothermophilic archaeon.
Nature, 478, 2011
|
|
2VAE
 
 | | Fast maturing red fluorescent protein, DsRed.T4 | | Descriptor: | 1,2-ETHANEDIOL, RED FLUORESCENT PROTEIN | | Authors: | Strongin, D.E, Bevis, B, Khuong, N, Downing, M.E, Strack, R.L, Sundaram, K, Glick, B.S, Keenan, R.J. | | Deposit date: | 2007-08-31 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Rearrangements Near the Chromophore Influence the Maturation Speed and Brightness of Dsred Variants.
Protein Eng.Des.Sel., 20, 2007
|
|
2V4R
 
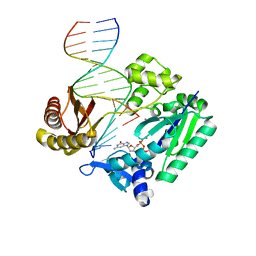 | | Non-productive complex of the Y-family DNA polymerase Dpo4 with dGTP skipping the M1dG adduct to pair with the next template cytosine | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*C)-3', 5'-D(*TP*CP*AP*CP*M1GP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Eoff, R.L, Stafford, J.B, Szekely, J, Rizzo, C.J, Egli, M, Guengerich, F.P, Marnett, L.J. | | Deposit date: | 2008-09-26 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Analysis of Sulfolobus Solfataricus Y-Family DNA Polymerase Dpo4-Catalyzed Bypass of the Malondialdehyde-Deoxyguanosine Adduct.
Biochemistry, 48, 2009
|
|
2VH5
 
 | | CRYSTAL STRUCTURE OF HRAS(G12V) - ANTI-RAS FV (disulfide free mutant) COMPLEX | | Descriptor: | ANTI-RAS FV HEAVY CHAIN, ANTI-RAS FV LIGHT CHAIN, GTPASE HRAS, ... | | Authors: | Tanaka, T, Williams, R.L, Rabbitts, T.H. | | Deposit date: | 2007-11-19 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Functional Intracellular Antibody Fragments Do not Require Invariant Intra-Domain Disulfide Bonds.
J.Mol.Biol., 376, 2008
|
|
2PFG
 
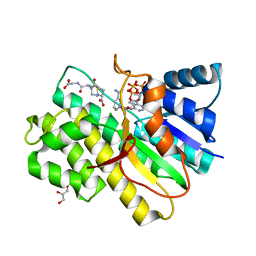 | | Crystal structure of human CBR1 in complex with BiGF2. | | Descriptor: | (5R,10S)-5-{[(CARBOXYMETHYL)AMINO]CARBONYL}-7-OXO-3-THIA-1,6-DIAZABICYCLO[4.4.1]UNDECANE-10-CARBOXYLIC ACID, CHLORIDE ION, Carbonyl reductase [NADPH] 1, ... | | Authors: | Rauh, D, Bateman, R.L, Shokat, K.M. | | Deposit date: | 2007-04-04 | | Release date: | 2007-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Glutathione traps formaldehyde by formation of a bicyclo[4.4.1]undecane adduct.
Org.Biomol.Chem., 5, 2007
|
|
3TOS
 
 | | Crystal Structure of CalS11, Calicheamicin Methyltransferase | | Descriptor: | 1,2-ETHANEDIOL, CalS11, GLUTAMIC ACID, ... | | Authors: | Chang, A, Aceti, D.J, Beebe, E.T, Makino, S.-I, Wrobel, R.L, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2011-09-06 | | Release date: | 2011-10-05 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of CalS11, Calicheamicin methyltransferase
To be Published
|
|
