4H7R
 
 | | Crystal structure of a parallel 4-helix coiled coil CC-Hex-II | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CC-Hex-II, GLYCEROL | | Authors: | Chi, B, Zaccai, N.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2012-09-20 | | Release date: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | TBA
To be Published
|
|
4HMZ
 
 | | Crystal Structure of ChmJ, a 3'-monoepimerase from Streptomyces bikiniensis in complex with dTDP-quinovose | | Descriptor: | 1,2-ETHANEDIOL, Putative 3-epimerase in D-allose pathway, [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl (2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-methyltetrahydro-2H-pyran-2-yl dihydrogen diphosphate | | Authors: | Holden, H.M, Kubiak, R.L. | | Deposit date: | 2012-10-18 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Studies on a 3'-Epimerase Involved in the Biosynthesis of dTDP-6-deoxy-d-allose.
Biochemistry, 51, 2012
|
|
5SXY
 
 | |
6HAL
 
 | | Human carbonmonoxy hemoglobin SFX dataset | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Doak, B, Gorel, A, Foucar, L, Barends, T.R.M, Gruenbein, M.L, Hilpert, M, Kloos, M, Nass Kovacs, G, Roome, C.M, Shoeman, R.L, Stricker, M, Tono, K, You, D, Ueda, K, Sherrell, D.A, Owen, R.L, Schlichting, I. | | Deposit date: | 2018-08-07 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallography on a chip - without the chip: sheet-on-sheet sandwich.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
2OQJ
 
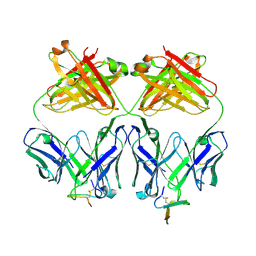 | | Crystal structure analysis of Fab 2G12 in complex with peptide 2G12.1 | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, peptide 2G12.1 (ACPPSHVLDMRSGTCLAAEGK) | | Authors: | Calarese, D.A, Stanfield, R.L, Menendez, A, Scott, J.K, Wilson, I.A. | | Deposit date: | 2007-01-31 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A peptide inhibitor of HIV-1 neutralizing antibody 2G12 is not a structural mimic of the natural carbohydrate epitope on gp120.
Faseb J., 22, 2008
|
|
5OQ4
 
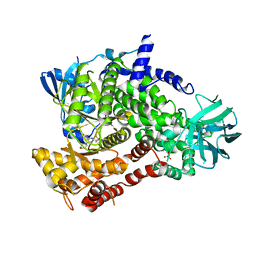 | | PQR309 - a Potent, Brain-Penetrant, Orally Bioavailable, pan-Class I PI3K/mTOR Inhibitor as Clinical Candidate in Oncology | | Descriptor: | 5-(4,6-dimorpholin-4-yl-1,3,5-triazin-2-yl)-4-(trifluoromethyl)pyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Williams, R.L, Zhang, X. | | Deposit date: | 2017-08-10 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 5-(4,6-Dimorpholino-1,3,5-triazin-2-yl)-4-(trifluoromethyl)pyridin-2-amine (PQR309), a Potent, Brain-Penetrant, Orally Bioavailable, Pan-Class I PI3K/mTOR Inhibitor as Clinical Candidate in Oncology.
J. Med. Chem., 60, 2017
|
|
6E3C
 
 | |
6QWG
 
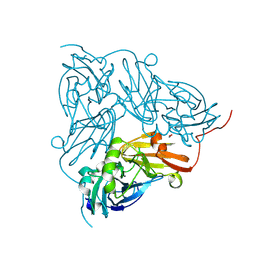 | | Serial Femtosecond Crystallography Structure of Cu Nitrite Reductase from Achromobacter cycloclastes: Nitrite complex at Room Temperature | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Ebrahim, A.E, Moreno-Chicano, T, Appleby, M.V, Worrall, J.W, Duyvesteyn, H.M.E, Strange, R.W, Beale, J, Axford, D, Sherrell, D.A, Sugimoto, H, Owada, S, Tono, K, Owen, R.L. | | Deposit date: | 2019-03-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-throughput structures of protein-ligand complexes at room temperature using serial femtosecond crystallography.
Iucrj, 6, 2019
|
|
2DOQ
 
 | | crystal structure of Sfi1p/Cdc31p complex | | Descriptor: | CALCIUM ION, Cell division control protein 31, SFI1p | | Authors: | Li, S, Sandercock, A.M, Conduit, P.T, Robinson, C.V, Williams, R.L, Kilmartin, J.V. | | Deposit date: | 2006-05-03 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication.
J.Cell Biol., 173, 2006
|
|
5T9W
 
 | | Discovery of a Potent Cyclophilin Inhibitor (Compound 5) based on Structural Simplification of Sanglifehrin A | | Descriptor: | 3-[(3-hydroxyphenyl)methyl]-6-(propan-2-yl)-19-oxa-1,4,7,25-tetraazabicyclo[19.3.1]pentacosa-13,15-diene-2,5,8,20-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Steadman, V, Pettit, P, Schmitz, U, Mackman, R.L, Schultz, B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Potent Cyclophilin Inhibitors Based on the Structural Simplification of Sanglifehrin A.
J. Med. Chem., 60, 2017
|
|
5T9U
 
 | | Discovery of a Potent Cyclophilin Inhibitor (Compound 3) based on Structural Simplification of Sanglifehrin A | | Descriptor: | 3-[(3-hydroxyphenyl)methyl]-10,12-dimethoxy-9,11-dimethyl-6-(propan-2-yl)-19-oxa-1,4,7,25-tetraazabicyclo[19.3.1]pentacosa-13,15-diene-2,5,8,20-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Steadman, V, Pettit, S, Schmitz, U, Mackman, R.L, Schultz, B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Discovery of Potent Cyclophilin Inhibitors Based on the Structural Simplification of Sanglifehrin A.
J. Med. Chem., 60, 2017
|
|
5T9Z
 
 | | Discovery of a Potent Cyclophilin Inhibitor (Compound 6) based on Structural Simplification of Sanglifehrin A | | Descriptor: | 11-[(3-hydroxyphenyl)methyl]-18-methoxy-17-methyl-14-(propan-2-yl)-3-oxa-9,12,15,28-tetraazatricyclo[21.3.1.1~5,9~]octacosa-1(27),21,23,25-tetraene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Steadman, V, Pettit, S, Schmitz, U, Mackman, R.L, Schultz, B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of Potent Cyclophilin Inhibitors Based on the Structural Simplification of Sanglifehrin A.
J. Med. Chem., 60, 2017
|
|
5T3S
 
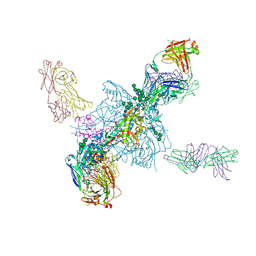 | | HIV gp140 trimer MD39-10MUTA in complex with Fabs PGT124 and 35022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2016-08-26 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | HIV Vaccine Design to Target Germline Precursors of Glycan-Dependent Broadly Neutralizing Antibodies.
Immunity, 45, 2016
|
|
5TA2
 
 | | Discovery of a Potent Cyclophilin Inhibitor (Compound 7) based on Structural Simplification of Sanglifehrin A | | Descriptor: | 11-[(3-hydroxyphenyl)methyl]-18-methoxy-2,17-dimethyl-14-(propan-2-yl)-3-oxa-9,12,15,28-tetraazatricyclo[21.3.1.1~5,9~]octacosa-1(27),21,23,25-tetraene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Steadman, V, Pettit, S, Schmitz, U, Mackman, R.L, Schultz, B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Discovery of Potent Cyclophilin Inhibitors Based on the Structural Simplification of Sanglifehrin A.
J. Med. Chem., 60, 2017
|
|
5TA4
 
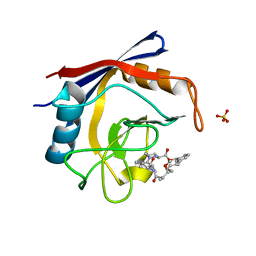 | | Discovery of a Potent Cyclophilin Inhibitor (Compound 8) based on Structural Simplification of Sanglifehrin A | | Descriptor: | 18-methoxy-2,11,17-trimethyl-14-(propan-2-yl)-3-oxa-9,12,15,28-tetraazatricyclo[21.3.1.1~5,9~]octacosa-1(27),21,23,25-tetraene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A, SULFATE ION | | Authors: | Appleby, T.C, Steadman, V, Pettit, S, Schmitz, U, Mackman, R.L, Schultz, B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Potent Cyclophilin Inhibitors Based on the Structural Simplification of Sanglifehrin A.
J. Med. Chem., 60, 2017
|
|
8B3S
 
 | | Structure of YjbA in complex with ClpC N-terminal Domain | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC / Negative regulator of tic competence clcC/mecB, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Evans, N.J, Isaacson, R.L, Camp, A.H, Collins, M.K. | | Deposit date: | 2022-09-16 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of YjbA in complex with ClpC N-terminal Domain
To Be Published
|
|
8C78
 
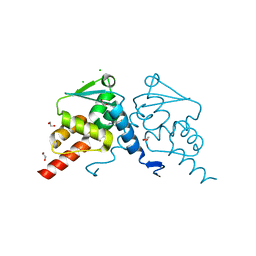 | | Crystal structure of human BCL6 BTB domain in complex with compound CCT374705 | | Descriptor: | (2~{S})-10-[(3-chloranyl-2-fluoranyl-pyridin-4-yl)amino]-2-cyclopropyl-3,3-bis(fluoranyl)-7-methyl-2,4-dihydro-1~{H}-[1,4]oxazepino[2,3-c]quinolin-6-one, 1,2-ETHANEDIOL, B-cell lymphoma 6 protein, ... | | Authors: | Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2023-01-13 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of an In Vivo Chemical Probe for BCL6 Inhibition by Optimization of Tricyclic Quinolinones.
J.Med.Chem., 66, 2023
|
|
4M92
 
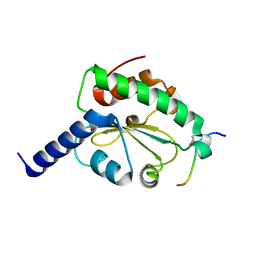 | | Crystal structure of hN33/Tusc3-peptide 2 | | Descriptor: | Interleukin-1 receptor accessory protein-like 1, Tumor suppressor candidate 3 | | Authors: | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
4M91
 
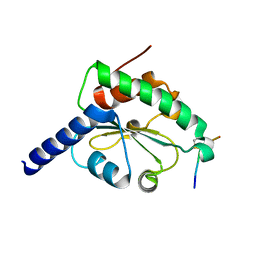 | | crystal structure of hN33/Tusc3-peptide 1 | | Descriptor: | Protein cereblon, Tumor suppressor candidate 3 | | Authors: | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
5JHB
 
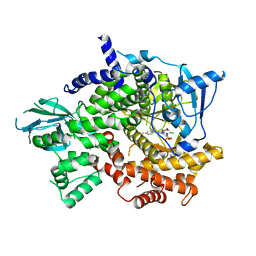 | | Structure of Phosphoinositide 3-kinase gamma (PI3K) bound to the potent inhibitor PIKin3 | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION, [1-{4-[6-amino-4-(trifluoromethyl)pyridin-3-yl]-6-(morpholin-4-yl)-1,3,5-triazin-2-yl}-3-(chloromethyl)azetidin-3-yl]methanol | | Authors: | Burke, J.E, Inglis, A.J, Williams, R.L. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Deconvolution of Buparlisib's mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention.
Nat Commun, 8, 2017
|
|
3NN8
 
 | |
5J74
 
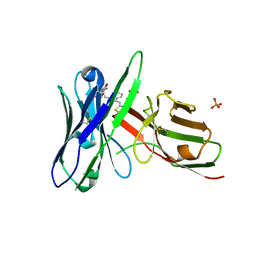 | | Fluorogen activating protein AM2.2 in complex with TO1-2p | | Descriptor: | 1-(17-amino-5,8-dioxo-12,15-dioxa-4,9-diazaheptadecan-1-yl)-4-{[3-(3-sulfopropyl)-1,3-benzothiazol-3-ium-2-yl]methyl}quinolin-1-ium, PHOSPHATE ION, scFv AM2.2 | | Authors: | Stanfield, R.L, Wilson, I.A, Wu, Y. | | Deposit date: | 2016-04-05 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Small-Molecule Nonfluorescent Inhibitors of Fluorogen-Fluorogen Activating Protein Binding Pair.
J Biomol Screen, 21, 2016
|
|
5J8D
 
 | | Structure of nitroreductase from E. cloacae complexed with nicotinic acid adenine dinucleotide | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINIC ACID ADENINE DINUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Haynes, C.A, Koder, R.L, Miller, A.F, Rodgers, D.W. | | Deposit date: | 2016-04-07 | | Release date: | 2017-05-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism-Informed Refinement Reveals Altered Substrate-Binding Mode for Catalytically Competent Nitroreductase.
Structure, 25, 2017
|
|
4M43
 
 | |
3NZP
 
 | | Crystal Structure of the Biosynthetic Arginine decarboxylase SpeA from Campylobacter jejuni, Northeast Structural Genomics Consortium Target BR53 | | Descriptor: | Arginine decarboxylase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-07-16 | | Release date: | 2010-09-01 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of bacterial biosynthetic arginine decarboxylases.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
