7BLG
 
 | | Structure of CBM BT3015C from Bacteroides thetaiotaomicron in complex with galactose | | Descriptor: | CALCIUM ION, beta-D-galactopyranose, family 32 carbohydrate-binding module from Bacteroides thetaiotaomicron | | Authors: | Costa, R.L. | | Deposit date: | 2021-01-18 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural basis for mucin-type O-glycan recognition by proteins of a Bacteroides thetaiotaomicron polysaccharide utilization loci
To Be Published
|
|
1FQ6
 
 | | X-RAY STRUCTURE OF GLYCOL INHIBITOR PD-133,450 BOUND TO SACCHAROPEPSIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(1S)-2-{[(2S,3R,4S)-1-cyclohexyl-3,4-dihydroxy-6-methylheptan-2-yl]amino}-1-(ethylsulfanyl)-2-oxoethyl]-Nalpha-(morp holin-4-ylsulfonyl)-L-phenylalaninamide, SACCHAROPEPSIN, ... | | Authors: | Cronin, N.B, Badasso, M.O, Tickle, I.J, Dreyer, T, Hoover, D.J, Rosati, R.L, Humblet, C.C, Lunney, E.A, Cooper, J.B. | | Deposit date: | 2000-09-03 | | Release date: | 2000-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray structures of five renin inhibitors bound to saccharopepsin: exploration of active-site specificity.
J.Mol.Biol., 303, 2000
|
|
7BLL
 
 | |
7BLH
 
 | |
7BLJ
 
 | |
6MUB
 
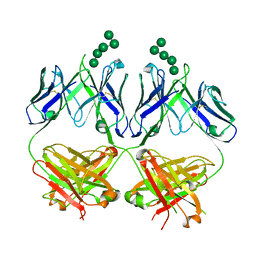 | | Anti-HIV-1 Fab 2G12 + Man5 re-refinement | | Descriptor: | Fab 2G12, heavy chain, light chain, ... | | Authors: | Wilson, I.A, Calarese, D.A, Stanfield, R.L. | | Deposit date: | 2018-10-22 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6N2X
 
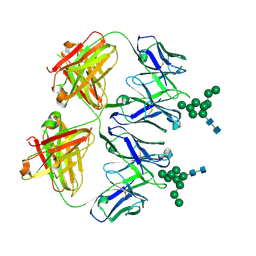 | | Anti-HIV-1 Fab 2G12 + Man9 re-refinement | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Calarese, D.A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-11-14 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Antibody domain exchange is an immunological solution to carbohydrate cluster recognition.
Science, 300, 2003
|
|
6MIE
 
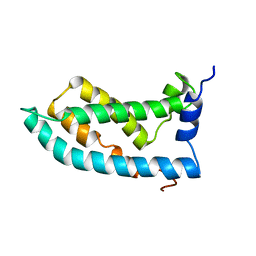 | | Solution NMR structure of the KCNQ1 voltage-sensing domain | | Descriptor: | Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Taylor, K.C, Kuenze, G, Smith, J.A, Meiler, J, McFeeters, R.L, Sanders, C.R. | | Deposit date: | 2018-09-19 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and physiological function of the human KCNQ1 channel voltage sensor intermediate state.
Elife, 9, 2020
|
|
6N32
 
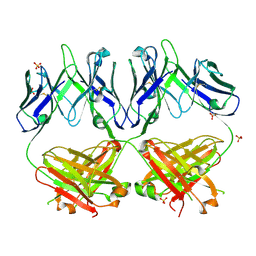 | | Anti-HIV-1 Fab 2G12 re-refinement | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, SULFATE ION | | Authors: | Calarese, D.A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-11-14 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antibody domain exchange is an immunological solution to carbohydrate cluster recognition.
Science, 300, 2003
|
|
1FQ5
 
 | | X-ray structure of a cyclic statine inhibitor PD-129,541 bound to yeast proteinase A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(5S,9S,10S,13S)-9-hydroxy-5,10-bis(2-methylpropyl)-4,7,12,16-tetraoxo-3,6,11,17-tetraazabicyclo[17.3.1]tricosa-1(23),19,21-trien-13-yl]-3-(naphthalen-1-yl)-2-(naphthalen-1-ylmethyl)propanamide, SACCHAROPEPSIN, ... | | Authors: | Cronin, N.B, Badasso, M.O, Tickle, I.J, Dreyer, T, Hoover, D.J, Rosati, R.L, Humblet, C.C, Lunney, E.A, Cooper, J.B. | | Deposit date: | 2000-09-03 | | Release date: | 2000-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of five renin inhibitors bound to saccharopepsin: exploration of active-site specificity.
J.Mol.Biol., 303, 2000
|
|
6MNF
 
 | | Anti-HIV-1 Fab 2G12 + Man8 re-refinement | | Descriptor: | Fab 2G12, light chain, Fab 2g12, ... | | Authors: | Calarese, D.A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.758 Å) | | Cite: | Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
1A3A
 
 | | CRYSTAL STRUCTURE OF IIA MANNITOL FROM ESCHERICHIA COLI | | Descriptor: | MANNITOL-SPECIFIC EII | | Authors: | Van Montfort, R.L.M, Pijning, T, Kalk, K.H, Hangyi, I, Kouwijzer, M.L.C.E, Robillard, G.T, Dijkstra, B.W. | | Deposit date: | 1998-01-19 | | Release date: | 1998-08-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the Escherichia coli phosphotransferase IIAmannitol reveals a novel fold with two conformations of the active site.
Structure, 6, 1998
|
|
6M87
 
 | | Fab 10A6 in complex with MPTS | | Descriptor: | 8-methoxypyrene-1,3,6-trisulfonic acid, Fab 10A6 heavy chain, Fab 10A6 light chain, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-08-21 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure and Dynamics of Stacking Interactions in an Antibody Binding Site.
Biochemistry, 58, 2019
|
|
6M94
 
 | | Monophosphorylated pSer33 b-Catenin peptide bound to b-TrCP/Skp1 Complex | | Descriptor: | Catenin beta-1, DIMETHYL SULFOXIDE, F-box/WD repeat-containing protein 1A, ... | | Authors: | Simonetta, K.R, Clifton, M.C, Walter, R.L, Ranieri, G.M, Carter, J.J. | | Deposit date: | 2018-08-22 | | Release date: | 2019-04-03 | | Last modified: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Prospective discovery of small molecule enhancers of an E3 ligase-substrate interaction.
Nat Commun, 10, 2019
|
|
1HYO
 
 | | CRYSTAL STRUCTURE OF FUMARYLACETOACETATE HYDROLASE COMPLEXED WITH 4-(HYDROXYMETHYLPHOSPHINOYL)-3-OXO-BUTANOIC ACID | | Descriptor: | 4-[HYDROXY-[METHYL-PHOSPHINOYL]]-3-OXO-BUTANOIC ACID, ACETATE ION, CALCIUM ION, ... | | Authors: | Bateman, R.L, Bhanumoorthy, P, Witte, J.F, McClard, R.W, Grompe, M, Timm, D.E. | | Deposit date: | 2001-01-21 | | Release date: | 2001-02-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanistic inferences from the crystal structure of fumarylacetoacetate hydrolase with a bound phosphorus-based inhibitor.
J.Biol.Chem., 276, 2001
|
|
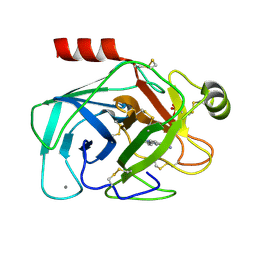
1GI6
 
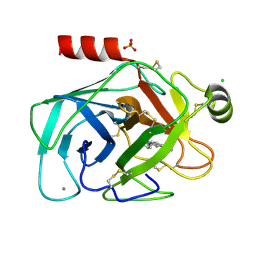 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-HYDROXY-PHENYL)-1H-INDOLE-5-CARBOXAMIDINE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GI1
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-HYDROXY-PHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GJC
 
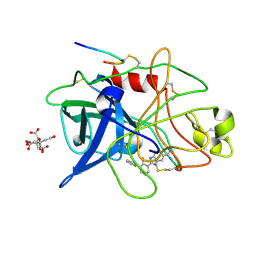 | | ENGINEERING INHIBITORS HIGHLY SELECTIVE FOR THE S1 SITES OF SER190 TRYPSIN-LIKE SERINE PROTEASE DRUG TARGETS | | Descriptor: | 2-(2-HYDROXY-BIPHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | Deposit date: | 2001-04-27 | | Release date: | 2002-04-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
6MEA
 
 | | Crystal structure of a Tylonycteris bat coronavirus HKU4 macrodomain in complex with adenosine diphosphate ribose (ADP-ribose) | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
1GHV
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-OXO-1,2-DIHYDRO-PYRIDIN-3-YL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, ACETYL HIRUDIN, SODIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GI9
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-HYDROXY-5-METHOXY-PHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GJB
 
 | | ENGINEERING INHIBITORS HIGHLY SELECTIVE FOR THE S1 SITES OF SER190 TRYPSIN-LIKE SERINE PROTEASE DRUG TARGETS | | Descriptor: | 2-(2-HYDROXY-BIPHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | Deposit date: | 2001-04-27 | | Release date: | 2002-04-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
6MU3
 
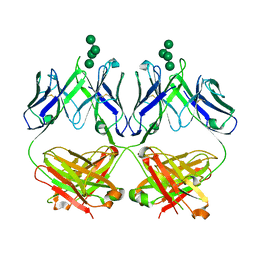 | | Anti-HIV-1 Fab 2G12 + Man7 re-refinement | | Descriptor: | Fab 2G12, heavy chain, light chain, ... | | Authors: | Wilson, I.A, Calarese, D.A, Stanfield, R.L. | | Deposit date: | 2018-10-22 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
1I0Z
 
 | | HUMAN HEART L-LACTATE DEHYDROGENASE H CHAIN, TERNARY COMPLEX WITH NADH AND OXAMATE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-LACTATE DEHYDROGENASE H CHAIN, OXAMIC ACID | | Authors: | Read, J.A, Winter, V.J, Eszes, C.M, Sessions, R.B, Brady, R.L. | | Deposit date: | 2001-01-30 | | Release date: | 2001-03-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for altered activity of M- and H-isozyme forms of human lactate dehydrogenase.
Proteins, 43, 2001
|
|
1I4E
 
 | | CRYSTAL STRUCTURE OF THE CASPASE-8/P35 COMPLEX | | Descriptor: | Caspase-8, Early 35 kDa protein | | Authors: | Xu, G, Cirilli, M, Huang, Y, Rich, R.L, Myszka, D.G, Wu, H. | | Deposit date: | 2001-02-20 | | Release date: | 2001-03-28 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Covalent inhibition revealed by the crystal structure of the caspase-8/p35 complex.
Nature, 410, 2001
|
|
