3NW8
 
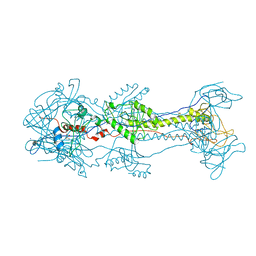 | | Glycoprotein B from Herpes simplex virus type 1, Y179S mutant, high-pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Stampfer, S.D, Lou, H, Cohen, G.H, Eisenberg, R.J, Heldwein, E.E. | | Deposit date: | 2010-07-09 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75915 Å) | | Cite: | Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1.
J.Virol., 84, 2010
|
|
1A39
 
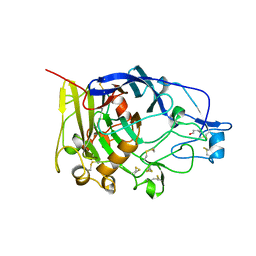 | | HUMICOLA INSOLENS ENDOCELLULASE EGI S37W, P39W DOUBLE-MUTANT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Davies, G.J, Ducros, V, Lewis, R.J, Borchert, T.V, Schulein, M. | | Deposit date: | 1998-01-28 | | Release date: | 1999-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oligosaccharide specificity of a family 7 endoglucanase: insertion of potential sugar-binding subsites.
J.Biotechnol., 57, 1997
|
|
5UDN
 
 | |
5UF1
 
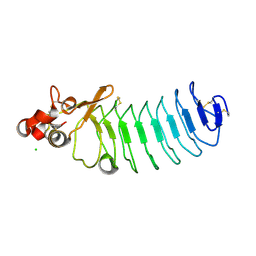 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) O13 in complex with H-trisaccharide | | Descriptor: | CHLORIDE ION, O13, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gunn, R.J, Collins, B.C, McKitrick, T.R, Cummings, R.D, Cooper, M.D, Herrin, B.R, Wilson, I.A. | | Deposit date: | 2017-01-03 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates.
Structure, 25, 2017
|
|
5UFD
 
 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) RBC36 (Apo) | | Descriptor: | MAGNESIUM ION, RBC36 | | Authors: | Collins, B.C, Gunn, R.J, McKitrick, T.R, Herrin, B.R, Cummings, R.D, Cooper, M.D, Wilson, I.A. | | Deposit date: | 2017-01-04 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates.
Structure, 25, 2017
|
|
8PDD
 
 | | Thioredoxin glutathione reductase of Schistosoma mansoni at 1.25A resolution. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ribeiro, L, Montoya, B.O, Moreira-Filho, J.T, Bowyer, S, Verma, A, Neves, B.J, Owens, R.J, Andrade, C.H, Silva-Jr, F.P, Furnham, N. | | Deposit date: | 2023-06-12 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Fragment library screening by X-ray crystallography and binding site analysis on thioredoxin glutathione reductase of Schistosoma mansoni.
Sci Rep, 14, 2024
|
|
1ALE
 
 | |
3OKN
 
 | | Crystal structure of S25-39 in complex with Kdo(2.4)Kdo(2.4)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain, ... | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-25 | | Release date: | 2011-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
5UFC
 
 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) Tn4-22 with H-trisaccharide bound | | Descriptor: | Tn4-22, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Collins, B.C, Gunn, R.J, McKitrick, T.R, Cummings, R.D, Cooper, M.D, Herrin, B.R, Wilson, I.A. | | Deposit date: | 2017-01-04 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates.
Structure, 25, 2017
|
|
2L1Q
 
 | |
5UDM
 
 | |
3OKD
 
 | | Crystal structure of S25-39 in complex with Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, DI(HYDROXYETHYL)ETHER, S25-39 Fab (IgG1k) heavy chain, ... | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-24 | | Release date: | 2011-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
1A95
 
 | | XPRTASE FROM E. COLI COMPLEXED WITH MG:CPRPP AND GUANINE | | Descriptor: | 1-ALPHA-PYROPHOSPHORYL-2-ALPHA,3-ALPHA-DIHYDROXY-4-BETA-CYCLOPENTANE-METHANOL-5-PHOSPHATE, BORIC ACID, GUANINE, ... | | Authors: | Vos, S, Parry, R.J, Burns, M.R, De Jersey, J, Martin, J.L. | | Deposit date: | 1998-04-16 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of free and complexed forms of Escherichia coli xanthine-guanine phosphoribosyltransferase.
J.Mol.Biol., 282, 1998
|
|
8OWT
 
 | | SARS-CoV-2 spike RBD with A8 and H3 nanobodies bound | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody A8, ... | | Authors: | Mikolajek, H, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
5UE5
 
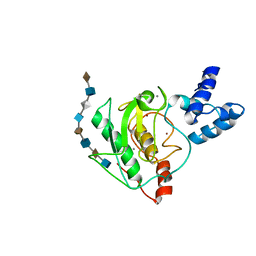 | | proMMP-7 with heparin octasaccharide bound to the catalytic domain | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Matrilysin, ... | | Authors: | Fulcher, Y.G, Prior, S.H, Linhardt, R.J, Van Doren, S.R. | | Deposit date: | 2016-12-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Glycan Activation of a Sheddase: Electrostatic Recognition between Heparin and proMMP-7.
Structure, 25, 2017
|
|
8OWW
 
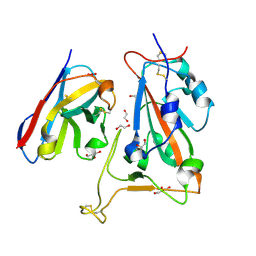 | | B5-5 nanobody bound to SARS-CoV-2 spike RBD (Wuhan) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, B5-5 nanobody, ... | | Authors: | Cornish, K.A.S, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
1A0M
 
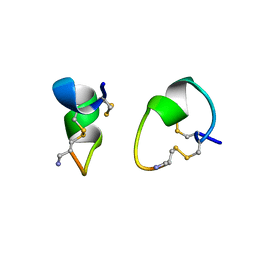 | | 1.1 ANGSTROM CRYSTAL STRUCTURE OF A-CONOTOXIN [TYR15]-EPI | | Descriptor: | ALPHA-CONOTOXIN [TYR15]-EPI | | Authors: | Hu, S.-H, Loughnan, M, Miller, R, Weeks, C.M, Blessing, R.H, Alewood, P.F, Lewis, R.J, Martin, J.L. | | Deposit date: | 1997-12-03 | | Release date: | 1999-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1 A resolution crystal structure of [Tyr15]EpI, a novel alpha-conotoxin from Conus episcopatus, solved by direct methods.
Biochemistry, 37, 1998
|
|
5UHY
 
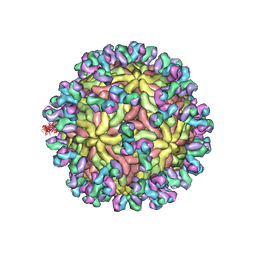 | | A Human Antibody Against Zika Virus Crosslinks the E Protein to Prevent Infection | | Descriptor: | ZV67 Fab chain 1, ZV67 Fab chain 2, envelope protein | | Authors: | Hasan, S.S, Miller, A, Sapparapu, G, Fernandez, E, Klose, T, Long, F, Fokine, A, Porta, J.C, Jiang, W, Diamond, M.S, Crowe Jr, J.E, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2017-01-12 | | Release date: | 2017-03-29 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | A human antibody against Zika virus crosslinks the E protein to prevent infection.
Nat Commun, 8, 2017
|
|
8OWV
 
 | | H6 and F2 nanobodies bound to SARS-CoV-2 spike RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2, GLYCEROL, ... | | Authors: | Mikolajek, H, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
5UE2
 
 | | proMMP-7 with heparin octasaccharide bridging between domains | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Matrilysin, ... | | Authors: | Fulcher, Y.G, Prior, S.H, Linhardt, R.J, Van Doren, S.R. | | Deposit date: | 2016-12-29 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Glycan Activation of a Sheddase: Electrostatic Recognition between Heparin and proMMP-7.
Structure, 25, 2017
|
|
8OYU
 
 | | Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '2 up 1 down' RBD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H6 nanobody, ... | | Authors: | Weckener, M, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
1A4O
 
 | | 14-3-3 PROTEIN ZETA ISOFORM | | Descriptor: | 14-3-3 PROTEIN ZETA | | Authors: | Liu, D, Bienkowska, J, Petosa, C, Collier, R.J, Fu, H, Liddington, R.C. | | Deposit date: | 1998-02-01 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the zeta isoform of the 14-3-3 protein.
Nature, 376, 1995
|
|
2L85
 
 | | Solution NMR structures of CBP bromodomain with small molecule of HBS | | Descriptor: | 4-[(E)-(4-hydroxyphenyl)diazenyl]benzenesulfonic acid, CREB-binding protein | | Authors: | Borah, J.C, Mujtaba, S, Karakikes, I, Zeng, L, Muller, M, Patel, J, Moshkina, N, Morohashi, K, Zhang, W, Gerona-Navarro, G, Hajjar, R.J, Zhou, M. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule Binding to the Coactivator CREB-Binding Protein Blocks Apoptosis in Cardiomyocytes.
Chem.Biol., 18, 2011
|
|
2LP8
 
 | | SOLUTION STRUCTURE OF AN APOPTOSIS ACTIVATING PHOTOSWITCHABLE BAK PEPTIDE BOUND to BCL-XL | | Descriptor: | 3,3'-(E)-diazene-1,2-diylbis{6-[(chloroacetyl)amino]benzenesulfonic acid}, Bcl-2 homologous antagonist/killer, Bcl-2-like protein 1 | | Authors: | Wysoczanski, P, Mart, R.J, Loveridge, J.E, Williams, C, Whittaker, S.B.-M, Crump, M.P, Allemann, R.K. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-18 | | Last modified: | 2021-08-18 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a Photoswitchable Apoptosis Activating Bak Peptide Bound to Bcl-x(L).
J.Am.Chem.Soc., 134, 2012
|
|
1AJY
 
 | | STRUCTURE AND MOBILITY OF THE PUT3 DIMER: A DNA PINCER, NMR, 13 STRUCTURES | | Descriptor: | PUT3, ZINC ION | | Authors: | Walters, K.J, Dayie, K.T, Reece, R.J, Ptashne, M, Wagner, G. | | Deposit date: | 1997-05-12 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and mobility of the PUT3 dimer.
Nat.Struct.Biol., 4, 1997
|
|
