7XQV
 
 | | The complex of nanobody Rh57 binding to GTP-bound RhoA active form | | Descriptor: | ALANINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Zhang, Y.R, Liu, R, Ding, Y. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural insights into the binding of nanobody Rh57 to active RhoA-GTP.
Biochem.Biophys.Res.Commun., 616, 2022
|
|
7Y45
 
 | | Cryo-EM structure of the Na+,K+-ATPase in the E2.2K+ state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-06-14 | | Release date: | 2022-07-13 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-electron microscopy of Na + ,K + -ATPase reveals how the extracellular gate locks in the E2·2K + state.
Febs Lett., 596, 2022
|
|
2OYS
 
 | | Crystal Structure of SP1951 protein from Streptococcus pneumoniae in complex with FMN, Northeast Structural Genomics Target SpR27 | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Hypothetical protein SP1951 | | Authors: | Forouhar, F, Lee, I, Vorobiev, S.M, Janjua, H, Satterwhite, R, Liu, J, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional insights from structural genomics.
J.Struct.Funct.Genom., 8, 2007
|
|
7Y46
 
 | | Cryo-EM structure of the Na+,K+-ATPase in the E2.2K+ state after addition of ATP | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-06-14 | | Release date: | 2022-07-13 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Cryo-electron microscopy of Na + ,K + -ATPase reveals how the extracellular gate locks in the E2·2K + state.
Febs Lett., 596, 2022
|
|
2W2E
 
 | | 1.15 Angstrom crystal structure of P.pastoris aquaporin, Aqy1, in a closed conformation at pH 3.5 | | Descriptor: | AQUAPORIN PIP2-7 7, CHLORIDE ION, octyl beta-D-glucopyranoside | | Authors: | Fischer, G, Kosinska-Eriksson, U, Aponte-Santamaria, C, Palmgren, M, Geijer, C, Hedfalk, K, Hohmann, S, de Groot, B.L, Neutze, R, Lindkvist-Petersson, K. | | Deposit date: | 2008-10-29 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of a Yeast Aquaporin at 1.15 A Reveals a Novel Gating Mechanism.1.15 A
Plos Biol., 7, 2009
|
|
2W39
 
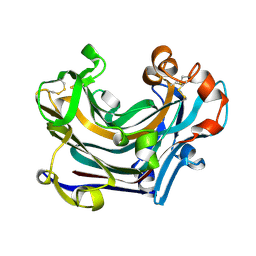 | | Glc(beta-1-3)Glc disaccharide in -1 and -2 sites of Laminarinase 16A from Phanerochaete chrysosporium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-glucono-1,5-lactone, PUTATIVE LAMINARINASE, ... | | Authors: | Vasur, J, Kawai, R, Andersson, E, Igarashi, K, Sandgren, M, Samejima, M, Stahlberg, J. | | Deposit date: | 2008-11-07 | | Release date: | 2009-07-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-Ray Crystal Structures of Phanerochaete Chrysosporium Laminarinase 16A in Complex with Products from Lichenin and Laminarin Hydrolysis
FEBS J., 276, 2009
|
|
2W62
 
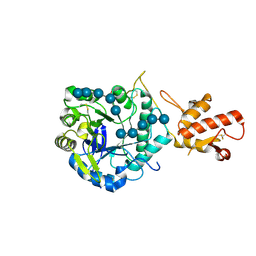 | | Saccharomyces cerevisiae Gas2p in complex with laminaripentaose | | Descriptor: | 1,4-BUTANEDIOL, GLYCOLIPID-ANCHORED SURFACE PROTEIN 2, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Schuettelkopf, A.W, Hurtado-Guerrero, R, van Aalten, D.M.F. | | Deposit date: | 2008-12-16 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Mechanisms of Yeast Cell Wall Glucan Remodeling.
J.Biol.Chem., 284, 2009
|
|
7CSV
 
 | | Pseudomonas aeruginosa antitoxin HigA | | Descriptor: | HTH cro/C1-type domain-containing protein | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
1BIZ
 
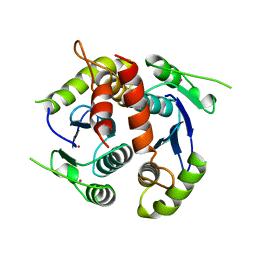 | | HIV-1 INTEGRASE CORE DOMAIN | | Descriptor: | CACODYLATE ION, HIV-1 INTEGRASE | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-21 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
2W5N
 
 | | Native structure of the GH93 alpha-L-arabinofuranosidase of Fusarium graminearum | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-L-ARABINOFURANOSIDASE, GLYCEROL, ... | | Authors: | Carapito, R, Imberty, A, Jeltsch, J.M, Byrns, S.C, Tam, P.H, Lowary, T.L, Varrot, A, Phalip, V. | | Deposit date: | 2008-12-11 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Basis of Arabinobio-Hydrolase Activity in Phytopathogenic Fungi. Crystal Structure and Catalytic Mechanism of Fusarium Graminearum Gh93 Exo-Alpha-L-Arabinanase.
J.Biol.Chem., 284, 2009
|
|
2WEH
 
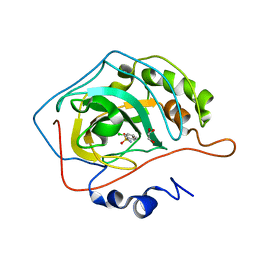 | | Thermodynamic Optimisation of Carbonic Anhydrase Fragment Inhibitors | | Descriptor: | 2-CHLOROBENZENESULFONAMIDE, CARBONIC ANHYDRASE 2, GLYCEROL, ... | | Authors: | Scott, A.D, Phillips, C, Alex, A, Bent, A, O'Brien, R, Damian, L, Jones, L.H. | | Deposit date: | 2009-03-31 | | Release date: | 2009-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Thermodynamic Optimisation in Drug Discovery: A Case Study Using Carbonic Anhydrase Inhibitors.
Chemmedchem, 4, 2009
|
|
2WDT
 
 | | Crystal structure of Plasmodium falciparum UCHL3 in complex with the suicide inhibitor UbVME | | Descriptor: | CHLORIDE ION, UBIQUITIN, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE L3 | | Authors: | Weihofen, W.A, Artavanis-Tsakonas, K, Gaudet, R, Ploegh, H.L. | | Deposit date: | 2009-03-26 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization and Structural Studies of the Plasmodium Falciparum Ubiquitin and Nedd8 Hydrolase Uchl3.
J.Biol.Chem., 285, 2010
|
|
2XH1
 
 | | Crystal structure of human KAT II-inhibitor complex | | Descriptor: | (3S)-10-(4-AMINOPIPERAZIN-1-YL)-9-FLUORO-7-HYDROXY-3-METHYL-2,3-DIHYDRO-8H-[1,4]OXAZINO[2,3,4-IJ]QUINOLINE-6-CARBOXYLATE, GLYCEROL, IODIDE ION, ... | | Authors: | Rossi, F, Casazza, V, Garavaglia, S, Sathyasaikumar, K.V, Schwarcz, R, Kojima, S.I, Okuwaki, K, Ono, S.I, Kajii, Y, Rizzi, M. | | Deposit date: | 2010-06-08 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure-Based Selective Targeting of the Pyridoxal 5'-Phsosphate Dependent Enzyme Kynurenine Aminotransferase II for Cognitive Enhancement
J.Med.Chem., 53, 2010
|
|
2XUM
 
 | | FACTOR INHIBITING HIF (FIH) Q239H MUTANT IN COMPLEX WITH ZN(II), NOG AND ASP-SUBSTRATE PEPTIDE (20-MER) | | Descriptor: | ASP-SUBSTRATE PEPTIDE 2, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR, N-OXALYLGLYCINE, ... | | Authors: | Chowdhury, R, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2010-10-19 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Asparagine and aspartate hydroxylation of the cytoskeletal ankyrin family is catalyzed by factor-inhibiting hypoxia-inducible factor.
J. Biol. Chem., 286, 2011
|
|
7B54
 
 | | VAR2CSA full ectodomain in present of plCS, DBL1-DBL4 | | Descriptor: | VAR2CSA in presence of plCS, DBl1-DBL4,Erythrocyte membrane protein 1 | | Authors: | Wang, K.T, Dagil, R, Gourdon, P.E, Salanti, A. | | Deposit date: | 2020-12-03 | | Release date: | 2021-06-02 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM reveals the architecture of placental malaria VAR2CSA and provides molecular insight into chondroitin sulfate binding.
Nat Commun, 12, 2021
|
|
2ODW
 
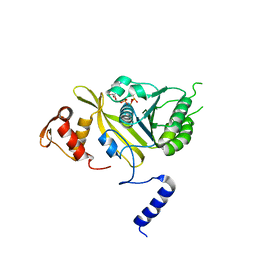 | | MSrecA-ATP-GAMA-S complex | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Protein recA | | Authors: | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2006-12-27 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
2XK3
 
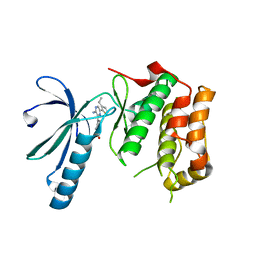 | | Structure of Nek2 bound to Aminopyrazine compound 35 | | Descriptor: | 4-[3-AMINO-6-(3-ETHYLTHIOPHEN-2-YL)PYRAZIN-2-YL]CYCLOHEXANE-1-CARBOXYLIC ACID, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2OIV
 
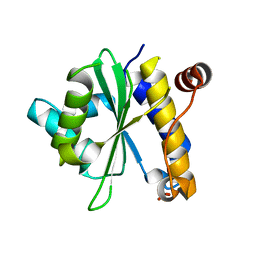 | | Structural Analysis of Xanthomonas XopD Provides Insights Into Substrate Specificity of Ubiquitin-like Protein Proteases | | Descriptor: | PHOSPHATE ION, Xanthomonas outer protein D | | Authors: | Chosed, R, Tomchick, D.R, Brautigam, C.A, Machius, M, Orth, K. | | Deposit date: | 2007-01-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases.
J.Biol.Chem., 282, 2007
|
|
2XI1
 
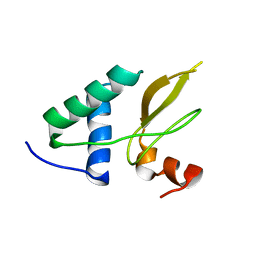 | | Crystal structure of the HIV-1 Nef sequenced from a patient's sample | | Descriptor: | NEF | | Authors: | Yadav, G.P, Singh, P, Gupta, S, Tripathi, A.K, Tripathi, R.K, Ramachandran, R. | | Deposit date: | 2010-06-25 | | Release date: | 2011-08-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A Novel Dimer-Tetramer Transition Captured by the Crystal Structure of the HIV-1 Nef.
Plos One, 6, 2011
|
|
7AFI
 
 | | Bacterial 30S ribosomal subunit assembly complex state C (body domain) | | Descriptor: | 16SrRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7AF3
 
 | | Bacterial 30S ribosomal subunit assembly complex state M (head domain) | | Descriptor: | 16S rRNA (head), 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S.R. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7AFN
 
 | | Bacterial 30S ribosomal subunit assembly complex state B (head domain) | | Descriptor: | 16SrRNA (head domain of the 30S ribosome), 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7AFD
 
 | | Bacterial 30S ribosomal subunit assembly complex state A (head domain) | | Descriptor: | 16SrRNA of the head domain (residue C931 to G1386), 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
2XO8
 
 | | Crystal Structure of Myosin-2 in Complex with Tribromodichloropseudilin | | Descriptor: | 2,4-DICHLORO-6-(3,4,5-TRIBROMO-1H-PYRROL-2-YL)PHENOL, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Preller, M, Chinthalapudi, K, Martin, R, Knoelker, H.J, Manstein, D.J. | | Deposit date: | 2010-08-10 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of Myosin ATPase Activity by Halogenated Pseudilins: A Structure-Activity Study.
J.Med.Chem., 54, 2011
|
|
2XRF
 
 | | Crystal structure of human uridine phosphorylase 2 | | Descriptor: | GLYCEROL, MAGNESIUM ION, URACIL, ... | | Authors: | Welin, M, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Kouznetsova, E, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Der Berg, S, Wahlberg, E, Weigelt, J, Nordlund, P. | | Deposit date: | 2010-09-14 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Human Uridine Phosphorylase 2
To be Published
|
|
