1KUT
 
 | | Structural Genomics, Protein TM1243, (SAICAR synthetase) | | Descriptor: | Phosphoribosylaminoimidazole-succinocarboxamide synthase | | Authors: | Zhang, R, Skarina, T, Beasley, S, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-22 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of SAICAR synthase from Thermotoga maritima at 2.2 angstroms reveals an unusual covalent dimer.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
7QTS
 
 | | GB1 in mammalian cells, 10 uM | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Gerez, J.A, Prymaczok, N.C, Kadavath, H, Gosh, D, Butikofer, M, Guntert, P, Riek, R. | | Deposit date: | 2022-01-15 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in human cells by in-cell NMR and a reporter system to optimize protein delivery or transexpression.
Commun Biol, 5, 2022
|
|
3OH8
 
 | | Crystal structure of the nucleoside-diphosphate sugar epimerase from Corynebacterium glutamicum. Northeast Structural Genomics Consortium Target CgR91 | | Descriptor: | Nucleoside-diphosphate sugar epimerase (SulA family) | | Authors: | Vorobiev, S, Lew, S, Kuzin, A, Mao, M, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-01 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Crystal structure of the nucleoside-diphosphate sugar epimerase from Corynebacterium glutamicum. Northeast Structural Genomics Consortium Target CgR91.
To be Published
|
|
3OF0
 
 | |
7QNZ
 
 | | human Lig1-DNA-PCNA complex reconstituted in absence of ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA ligase 1, Oligo13P, ... | | Authors: | Blair, K, Tehseen, M, Raducanu, V.S, Shahid, T, Lancey, C, Cruehet, R, Hamdan, S, De Biasio, A. | | Deposit date: | 2021-12-23 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.58 Å) | | Cite: | Mechanism of human Lig1 regulation by PCNA in Okazaki fragment sealing.
Nat Commun, 13, 2022
|
|
7QO1
 
 | | complex of DNA ligase I and FEN1 on PCNA and DNA | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA ligase 1, Flap endonuclease 1, ... | | Authors: | Blair, K, Tehseen, M, Raducanu, V.S, Shahid, T, Lancey, C, Cruehet, R, Hamdan, S, De Biasio, A. | | Deposit date: | 2021-12-23 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mechanism of human Lig1 regulation by PCNA in Okazaki fragment sealing.
Nat Commun, 13, 2022
|
|
4C89
 
 | | Crystal structure of carboxylesterase LpEst1 from Lactobacillus plantarum: high resolution model | | Descriptor: | ESTERASE, GLYCEROL, MALONATE ION | | Authors: | Alvarez, Y, Esteban-Torres, M, Cortes-Cabrera, A, Gago, F, Acebron, I, Benavente, R, Mardo, K, de-las-Rivas, B, Munoz, R, Mancheno, J.M. | | Deposit date: | 2013-09-30 | | Release date: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Esterase Lpest1 from Lactobacillus Plantarum: A Novel and Atypical Member of the Alpha Beta Hydrolase Superfamily of Enzymes
Plos One, 9, 2014
|
|
8R09
 
 | | Cryo-EM structure of the cross-exon pre-B+5'ss+ATPgammaS complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
4BZW
 
 | | Complete crystal structure of the carboxylesterase Cest-2923 (lp_2923) from Lactobacillus plantarum WCFS1 | | Descriptor: | ACETATE ION, LIPASE/ESTERASE, SULFATE ION, ... | | Authors: | Benavente, R, Esteban-Torres, M, Acebron, I, delasRivas, B, Munoz, R, Alvarez, Y, Mancheno, J.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Structure, Biochemical Characterization and Analysis of the Pleomorphism of Carboxylesterase Cest-2923 from Lactobacillus Plantarum Wcfs1
FEBS J., 280, 2013
|
|
4R2B
 
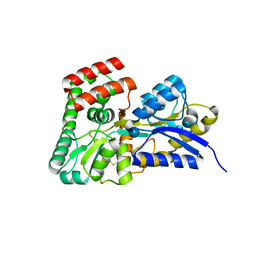 | | Crystal structure of sugar transporter Oant_3817 from Ochrobactrum anthropi, target EFI-510528, with bound glucose | | Descriptor: | Extracellular solute-binding protein family 1, alpha-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-11 | | Release date: | 2014-08-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure of Glucose Transporter Oant_3817 from Ochrobactrum Anthropi, Target EFI-510528
To be Published
|
|
8QOZ
 
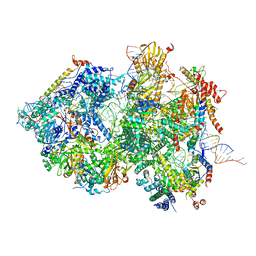 | | Cryo-EM Structure of Pre-B+5'ss+ATPgammaS Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss RNA oligo, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-09-29 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8QZS
 
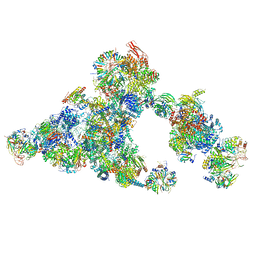 | | Cryo-EM structure of the cross-exon B-like complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Microfibrillar-associated protein 1, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-29 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
3OTL
 
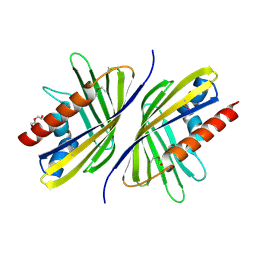 | | Three-dimensional Structure of the putative uncharacterized protein from Rhizobium leguminosarum at the resolution 1.9A, Northeast Structural Genomics Consortium Target RlR261 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Putative uncharacterized protein | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-09-13 | | Release date: | 2010-09-22 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Northeast Structural Genomics Consortium Target RlR261
To be Published
|
|
6GT3
 
 | | Crystal Structure of the A2A-StaR2-bRIL562 in complex with AZD4635 at 2.0A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 6-(2-chloranyl-6-methyl-pyridin-4-yl)-5-(4-fluorophenyl)-1,2,4-triazin-3-amine, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, ... | | Authors: | Borodovsky, A, Wang, Y, Deng, N, Ye, M, Stephen, T.L, Goodwin, K, Goodwin, R, Strittmatter, N, Shaw, J, Sachsenmeier, K, Clarke, J.D, Hay, C, Reimer, C, Andrews, S.P, Brown, G.A, Congreve, M, Cheng, R.K.Y, Dore, A.S, Mason, J.S, Marshall, F.H, Weir, M.P, Lyne, P, Woessner, R. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small molecule AZD4635 inhibitor of A2AR signaling rescues immune cell function including CD103+ dendritic cells enhancing anti-tumor immunity
J Immunother Cancer, 2020
|
|
4QK0
 
 | | Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 | | Descriptor: | GH127 beta-L-arabinofuranoside | | Authors: | Lansky, S, Salama, R, Dann, R, Shner, I, Manjasetty, B, Belrhali, H, Shoham, Y, Shoham, G. | | Deposit date: | 2014-06-05 | | Release date: | 2015-06-10 | | Method: | X-RAY DIFFRACTION (2.258 Å) | | Cite: | Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6
To be Published
|
|
7R8V
 
 | | Cryo-EM structure of the ADP state actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gong, R, Espinosa de los Reyes, S, Reynolds, M.J, Gurel, P, Alushin, G.M. | | Deposit date: | 2021-06-27 | | Release date: | 2021-07-28 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia.
Sci Adv, 8, 2022
|
|
7R91
 
 | | cryo-EM structure of the rigor state wild type myosin-15-F-actin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gong, R, Reynolds, M.J, Gurel, P, Alushin, G.M. | | Deposit date: | 2021-06-28 | | Release date: | 2021-07-28 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia.
Sci Adv, 8, 2022
|
|
7RB9
 
 | | Cryo-EM structure of the rigor state Jordan myosin-15-F-actin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gong, R, Reynolds, M.J, Gurel, P, Alushin, G.M. | | Deposit date: | 2021-07-05 | | Release date: | 2021-07-28 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia.
Sci Adv, 8, 2022
|
|
7RB8
 
 | | cryo-EM structure of the ADP state wild type myosin-15-F-actin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gong, R, Reynolds, M.J, Gurel, P, Alushin, G.M. | | Deposit date: | 2021-07-05 | | Release date: | 2021-07-28 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia.
Sci Adv, 8, 2022
|
|
8QXD
 
 | | Cryo-EM structure of the cross-exon pre-B complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, NHP2-like protein 1, N-terminally processed, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8QP8
 
 | | Cryo-EM Structure of Pre-B Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, INOSITOL HEXAKISPHOSPHATE, Pre-mRNA-processing factor 6, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-09-30 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8R0A
 
 | | Cryo-EM structure of the cross-exon pre-B+5'ss complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
4YT3
 
 | | CYP106A2 | | Descriptor: | ACETATE ION, Cytochrome P450(MEG), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | janocha, S, carius, y, bernhardt, r, lancaster, c.r.d. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of CYP106A2 in Substrate-Free and Substrate-Bound Form.
Chembiochem, 17, 2016
|
|
2VF0
 
 | | CRYSTAL STRUCTURE OF THE THYMIDYLATE SYNTHASE K48Q COMPLEXED WITH 5NO2DUMP AND BW1843U89 | | Descriptor: | 2'-DEOXY-5-NITROURIDINE 5'-MONOPHOSPHATE, S)-2-(5(((1,2-DIHYDRO-3-METHYL-1-OXOBENZO(F)QUINAZOLIN-9-YL)METHYL)AMINO)1-OXO-2-ISOINDOLINYL)GLUTARIC ACID, SULFATE ION, ... | | Authors: | Sotelo-Mundo, R.R, Arreola, R, Maley, F, Montfort, W.R. | | Deposit date: | 2007-10-27 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Role of an Invariant Lysine Residue in Folate Binding on Escherichia Coli Thymidylate Synthase: Calorimetric and Crystallographic Analysis of the K48Q Mutant.
Int.J.Biochem.Cell Biol., 40, 2008
|
|
8QPE
 
 | | Cryo-EM Structure of Pre-B-like Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, INOSITOL HEXAKISPHOSPHATE, Microfibrillar-associated protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-01 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
