5LC0
 
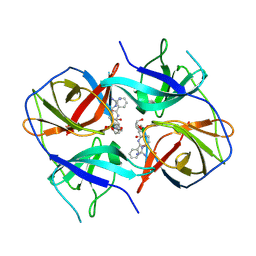 | | Crystal structure of Zika virus NS2B-NS3 protease in complex with a boronate inhibitor | | Descriptor: | N-((S)-3-(4-(aminomethyl)phenyl)-1-(((R)-4-guanidino-1-(5-hydroxy-1,3,2-dioxaborinan-2-yl)butyl)amino)-1-oxopropan-2-yl)benzamide, NS2B-NS3 protease,NS2B-NS3 protease | | Authors: | Lei, J, Hansen, G, Zhang, L.L, Hilgenfeld, R. | | Deposit date: | 2016-06-18 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Zika virus NS2B-NS3 protease in complex with a boronate inhibitor.
Science, 353, 2016
|
|
7KWX
 
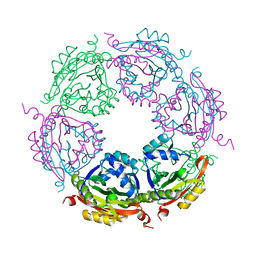 | | Spermidine N-acetyltransferase SpeG N152L mutant from Vibrio cholerae | | Descriptor: | Spermidine N(1)-acetyltransferase | | Authors: | Le, V.T.B, Tsimbalyuk, S, Lim, E.Q, Solis, A, Gawat, D, Boeck, P, Renolo, R, Forwood, J.K, Kuhn, M.L. | | Deposit date: | 2020-12-02 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The Vibrio cholerae SpeG Spermidine/Spermine N -Acetyltransferase Allosteric Loop and beta 6-beta 7 Structural Elements Are Critical for Kinetic Activity.
Front Mol Biosci, 8, 2021
|
|
5BMV
 
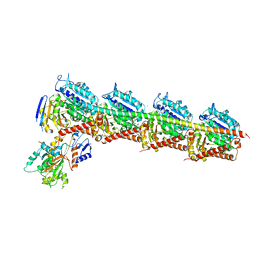 | | CRYSTAL STRUCTURE OF TUBULIN-STATHMIN-TTL-Vinblastine COMPLEX | | Descriptor: | (2ALPHA,2'BETA,3BETA,4ALPHA,5BETA)-VINCALEUKOBLASTINE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Chen, Q, Zhang, R. | | Deposit date: | 2015-05-23 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules.
Mol.Pharmacol., 89, 2016
|
|
7KWQ
 
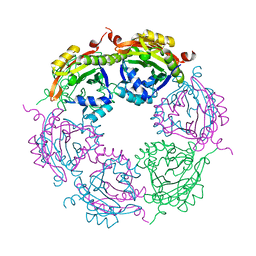 | | Spermidine N-acetyltransferase SpeG R149-K152 chimera from Vibrio cholerae and hSSAT | | Descriptor: | Spermidine N(1)-acetyltransferase | | Authors: | Le, V.T.B, Tsimbalyuk, S, Lim, E.Q, Solis, A, Gawat, D, Boeck, P, Renolo, R, Forwood, J.K, Kuhn, M.L. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Vibrio cholerae SpeG Spermidine/Spermine N -Acetyltransferase Allosteric Loop and beta 6-beta 7 Structural Elements Are Critical for Kinetic Activity.
Front Mol Biosci, 8, 2021
|
|
5B2P
 
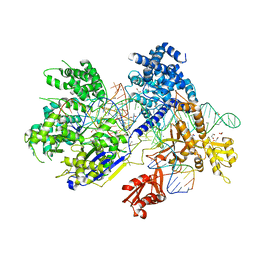 | | Crystal structure of Francisella novicida Cas9 in complex with sgRNA and target DNA (TGA PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Hirano, H, Nishimasu, H, Nakane, T, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-01 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Engineering of Francisella novicida Cas9
Cell, 164, 2016
|
|
4QD2
 
 | | Molecular basis for disruption of E-cadherin adhesion by botulinum neurotoxin A complex | | Descriptor: | CALCIUM ION, Cadherin-1, Hemagglutinin component HA17, ... | | Authors: | Lee, K, Zhong, X, Gu, S, Kruel, A, Dorner, M.B, Perry, K, Rummel, A, Dong, M, Jin, R. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for disruption of E-cadherin adhesion by botulinum neurotoxin A complex.
Science, 344, 2014
|
|
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | Descriptor: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KOA
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Shuvalova, L, Lavens, A, Henning, R, Maltseva, N, Rosas-Lemus, M, Kim, Y, Satchell, K.J.F, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography
To Be Published
|
|
2M0Z
 
 | | cis form of a photoswitchable PDZ domain crosslinked with an azobenzene derivative | | Descriptor: | 3,3'-(E)-diazene-1,2-diylbis{6-[(chloroacetyl)amino]benzenesulfonic acid}, Tyrosine-protein phosphatase non-receptor type 13 | | Authors: | Walser, R, Zerbe, O, Hamm, P. | | Deposit date: | 2012-11-09 | | Release date: | 2013-07-03 | | Last modified: | 2013-08-07 | | Method: | SOLUTION NMR | | Cite: | Kinetic response of a photoperturbed allosteric protein.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7KOJ
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder494 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]-5-{[(prop-2-en-1-yl)carbamoyl]amino}benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder494
to be published
|
|
2IMM
 
 | |
5BQU
 
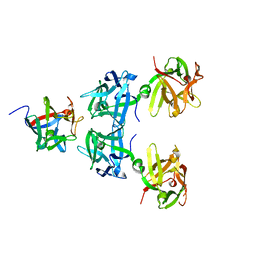 | | Crystal structure of HA17-HA33-Lactulose | | Descriptor: | HA-17, HA-33, beta-D-galactopyranose-(1-4)-beta-D-fructofuranose | | Authors: | Lee, K, Lam, K, Jin, R. | | Deposit date: | 2015-05-29 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Inhibiting oral intoxication of botulinum neurotoxin A complex by carbohydrate receptor mimics.
Toxicon, 107, 2015
|
|
5LZB
 
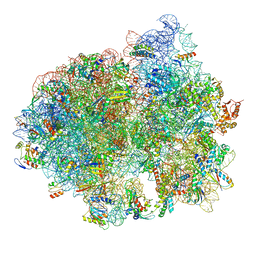 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the initial binding state (IB) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5BRZ
 
 | | MAGE-A3 reactive TCR in complex with MAGE-A3 in HLA-A1 | | Descriptor: | Beta-2-microglobulin, GLU-VAL-ASP-PRO-ILE-GLY-HIS-LEU-TYR, HLA class I histocompatibility antigen, ... | | Authors: | Raman, M.C.C, Rizkallah, P.J, Simmons, R, Donnellan, Z, Dukes, J, Bossi, G, LeProvost, G, Mahon, T, Hickman, E, LomaX, M, Oates, J, Hassan, N, Vuidepot, A, Sami, M, Cole, D.K, Jakobsen, B.K. | | Deposit date: | 2015-06-01 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Direct molecular mimicry enables off-target cardiovascular toxicity by an enhanced affinity TCR designed for cancer immunotherapy.
Sci Rep, 6, 2016
|
|
4QO1
 
 | | p53 DNA binding domain in complex with Nb139 | | Descriptor: | Cellular tumor antigen p53, Nb139 Nanobody against the DNA-binding domain of p53, ZINC ION | | Authors: | De Gieter, S, Bethuyne, J, Gettemans, J, Garcia-Pino, A, Loris, R. | | Deposit date: | 2014-06-19 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.924 Å) | | Cite: | A nanobody modulates the p53 transcriptional program without perturbing its functional architecture.
Nucleic Acids Res., 42, 2014
|
|
5M57
 
 | | Nek2 bound to arylaminopurine 6 | | Descriptor: | O6-CYCLOHEXYLMETHOXY-2-(4'-SULPHAMOYLANILINO) PURINE, Serine/threonine-protein kinase Nek2 | | Authors: | Bayliss, R. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of purine-based probes for selective Nek2 inhibition.
Oncotarget, 8, 2017
|
|
5M1W
 
 | | Structure of a stable G-hairpin | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*GP*GP*GP*TP*GP*TP*G)-3') | | Authors: | Gajarsky, M, Zivkovic, M.L, Stadlbauer, P, Pagano, B, Fiala, R, Amato, J, Tomaska, L, Sponer, J, Plavec, J, Trantirek, L. | | Deposit date: | 2016-10-11 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a Stable G-Hairpin.
J. Am. Chem. Soc., 139, 2017
|
|
5BQ1
 
 | | Capturing Carbon Dioxide in beta Carbonic Anhydrase | | Descriptor: | CARBON DIOXIDE, Carbonic anhydrase, ZINC ION | | Authors: | Aggarwal, M, Chua, T.K, Pinard, M.A, Szebenyi, D.M, McKenna, R. | | Deposit date: | 2015-05-28 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Carbon Dioxide "Trapped" in a beta-Carbonic Anhydrase.
Biochemistry, 54, 2015
|
|
4QKZ
 
 | | X-ray structure of the catalytic domain of MMP-8 with the inhibitor ML115 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Neutrophil collagenase, ... | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Tortorella, P. | | Deposit date: | 2014-06-10 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Bone-Seeking Matrix Metalloproteinase Inhibitors for the Treatment of Skeletal Malignancy.
Pharmaceuticals, 13, 2020
|
|
7YV3
 
 | |
7KRX
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441 inhibitor | | Descriptor: | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441
to be published
|
|
5BU4
 
 | | RIBONUCLEASE T1 COMPLEX WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Loris, R, Devos, S, Langhorst, U, Decanniere, K, Bouckaert, J, Maes, D, Transue, T.R, Steyaert, J. | | Deposit date: | 1998-09-15 | | Release date: | 1998-09-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Conserved water molecules in a large family of microbial ribonucleases.
Proteins, 36, 1999
|
|
7YV5
 
 | |
5LDL
 
 | | Myristoylated T41I/T78I mutant of M-PMV matrix protein | | Descriptor: | MYRISTIC ACID, myristoylated M-PMV matrix protein mutant | | Authors: | Kroupa, T, Hrabal, R. | | Deposit date: | 2016-06-27 | | Release date: | 2016-07-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Membrane Interactions of the Mason-Pfizer Monkey Virus Matrix Protein and Its Budding Deficient Mutants.
J.Mol.Biol., 428, 2016
|
|
5BR8
 
 | | Ambient-temperature crystal structure of 30S ribosomal subunit from Thermus thermophilus in complex with paromomycin | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Sierra, R.G, Gati, C, Laksmono, H, Dao, E.H, Gul, S, Fuller, F, Kern, J, Chatterjee, R, Ibrahim, M, Brewster, A, Young, I.D, Michels-Clark, T, Aquila, A, Mengning, L, Hunter, M.S, Koglin, J.E, Boutet, S, Junco, E.A, Hayes, B, Bogan, M.J, Hampton, C.Y, Puglisi, E.V, Sauter, N.K, Stan, C.A, Zouni, A, Yano, J, Yachandra, V.K, Soltis, S.M, Puglisi, J.D, DeMirci, H. | | Deposit date: | 2015-05-29 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ambient-temperature crystal structure of 30S ribosomal subunit from Thermus thermophilus in complex with paromomycin
To Be Published
|
|
