4WUT
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE | | Descriptor: | ABC transporter substrate binding protein (Ribose), CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-03 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE
To be published
|
|
8ETB
 
 | | the crystal structure of a rationally designed zinc sensor based on maltose binding protein - Zn binding conformation | | Descriptor: | ACETATE ION, ZINC ION, Zinc Sensor protein | | Authors: | Zhao, Z, Zhou, M, Zemerov, S.D, Marmorstein, R, Dmochowski, I.J. | | Deposit date: | 2022-10-16 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Rational design of a genetically encoded NMR zinc sensor.
Chem Sci, 14, 2023
|
|
8QQY
 
 | | Crystal structure of human Ephrin type-A receptor 2 (EPHA2) Kinase domain in complex with JG165 | | Descriptor: | 1,2-ETHANEDIOL, 6-(4-fluoranyl-3-methoxy-phenyl)-13$l^{6}-thia-2,4,8,12,19-pentazatricyclo[12.3.1.1^{3,7}]nonadeca-1(18),3,5,7(19),14,16-hexaene 13,13-dioxide, Ephrin type-A receptor 2 | | Authors: | Zhubi, R, Gerninghaus, J, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-06 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Ephrin type-A receptor 2 (EPHA2) Kinase domain in complex with JG165
To Be Published
|
|
7RQB
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site aminoacyl-tRNA analog ACC-PMN, and P-site MAI-tripeptidyl-tRNA analog ACCA-IAM at 2.45A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8PMJ
 
 | | Vanadate-trapped BSEP in nanodiscs | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Bile salt export pump, ... | | Authors: | Liu, H, Irobalieva, R.N, Kowal, J, Ni, D, Nosol, K, Bang-Sorensen, R, Lancien, L, Stahlberg, H, Stieger, B, Locher, K.P. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural basis of bile salt extrusion and small-molecule inhibition in human BSEP.
Nat Commun, 14, 2023
|
|
4X2C
 
 | | Clostridium difficile Fic protein_0569 mutant S31A, E35A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fic family protein putative filamentation induced by cAMP protein, GLYCEROL, ... | | Authors: | Jorgensen, R, Dedic, E. | | Deposit date: | 2014-11-26 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Clostridium difficile Fic_0569 S31A, E35A mutant at 1.8 Angstroms resolution
To Be Published
|
|
4X80
 
 | | Crystal Structure of murine 7B4 Fab monoclonal antibody against ADAMTS5 | | Descriptor: | IgG1 7B4 FAB Heavy chain, IgG1 7B4 FAB Light Chain | | Authors: | Larkin, J, Lohr, T.A, Elefante, L, Shearin, J, Matico, R, Su, J.-L, Xue, Y, Liu, F, Genell, C, Miller, R.E, Tran, P.B, Malfait, A.-M, Maier, C.C, Matheny, C.J. | | Deposit date: | 2014-12-09 | | Release date: | 2015-04-08 | | Last modified: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Translational development of an ADAMTS-5 antibody for osteoarthritis disease modification.
Osteoarthr. Cartil., 23, 2015
|
|
8PVB
 
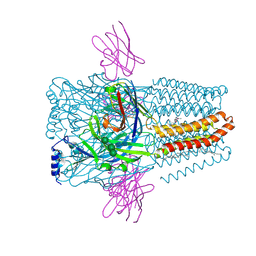 | | Structure of GABAAR determined by cryoEM at 100 keV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DECANE, ... | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PVI
 
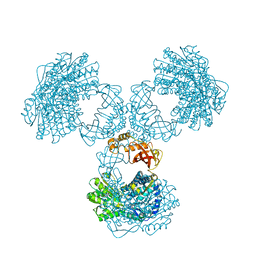 | | Structure of PaaZ determined by cryoEM at 100 keV | | Descriptor: | Bifunctional protein PaaZ | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4X8J
 
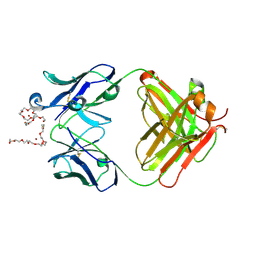 | | Crystal Structure of murine 12F4 Fab monoclonal antibody against ADAMTS5 | | Descriptor: | 12F4 FAB Heavy chain, 12F4 FAB Light chain, NONAETHYLENE GLYCOL, ... | | Authors: | Larkin, J, Lohr, T.A, Elefante, L, Shearin, J, Matico, R, Su, J.-L, Xue, Y, Liu, F, Genell, C, Miller, R.E, Tran, P.B, Malfait, A.-M, Maier, C.C, Matheny, C.J. | | Deposit date: | 2014-12-10 | | Release date: | 2015-04-08 | | Last modified: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Translational development of an ADAMTS-5 antibody for osteoarthritis disease modification.
Osteoarthr. Cartil., 23, 2015
|
|
8PVC
 
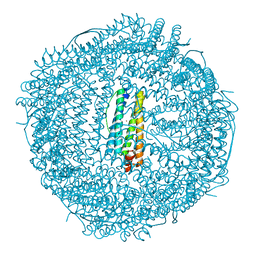 | | Structure of mouse heavy-chain apoferritin determined by cryoEM at 100 keV | | Descriptor: | FE (III) ION, Ferritin heavy chain, ZINC ION | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PVE
 
 | | Structure of AHIR determined by cryoEM at 100 keV | | Descriptor: | Ketol-acid reductoisomerase (NADP(+)) | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PVJ
 
 | | Structure of lumazine synthase determined by cryoEM at 100 keV | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PVF
 
 | | Structure of GAPDH determined by cryoEM at 100 keV | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PVD
 
 | | Structure of catalase determined by cryoEM at 100 keV | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6QIQ
 
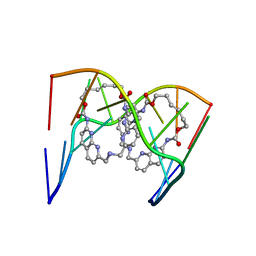 | | Crystal structure of seleno-derivative CAG repeats with synthetic CMBL3a compound | | Descriptor: | CMBL3a, RNA (5'-R(*GP*CP*AP*G)-D(P*(CSL))-R(P*AP*GP*C)-3') | | Authors: | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Micura, R, Nakatani, K. | | Deposit date: | 2019-01-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.519 Å) | | Cite: | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
4GO8
 
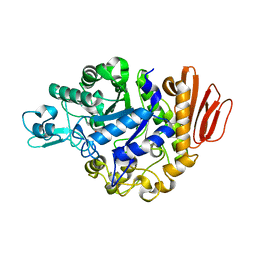 | | Crystal Structure of the TREHALULOSE SYNTHASE MUTB, MUTANT A258V, in complex with TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Sucrose isomerase | | Authors: | Lipski, A, Haser, R, Aghajari, N. | | Deposit date: | 2012-08-19 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of MUTB A258V mutant in complex with TRIS
To be Published, 2013
|
|
4X6Z
 
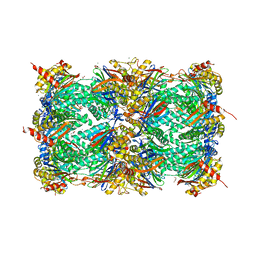 | | Yeast 20S proteasome in complex with PR-VI modulator | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Rostankowski, R, Witkowska, J, Borek, D, Otwinowski, Z, Jankowska, E. | | Deposit date: | 2014-12-09 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures revealed the common place of binding of low-molecular
mass activators with the 20S proteasome
To Be Published
|
|
8PMD
 
 | | Nucleotide-bound BSEP in nanodiscs | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bile salt export pump, MAGNESIUM ION | | Authors: | Liu, H, Irobalieva, R.N, Kowal, J, Ni, D, Nosol, K, Bang-Sorensen, R, Lancien, L, Stahlberg, H, Stieger, B, Locher, K.P. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of bile salt extrusion and small-molecule inhibition in human BSEP.
Nat Commun, 14, 2023
|
|
8PV9
 
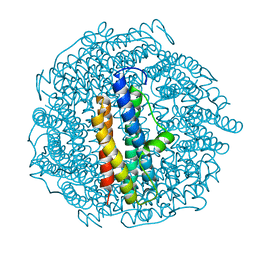 | | Structure of DPS determined by cryoEM at 100 keV | | Descriptor: | DNA protection during starvation protein | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PVH
 
 | | Structure of human apo ALDH1A1 determined by cryoEM at 100 keV | | Descriptor: | Aldehyde dehydrogenase 1A1, CHLORIDE ION | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4YM9
 
 | | Crystal structure of Porcine Pancreatic Elastase (PPE) in complex with the novel inhibitor JM102 | | Descriptor: | 2-ethyl-2-(hydroxymethyl)-N-(6-methylpyridin-3-yl)butanamide, ACETATE ION, Chymotrypsin-like elastase family member 1, ... | | Authors: | Hofbauer, S, Brito, J.A, Mulchande, J, Nogly, P, Pessanha, M, Moreira, R, Archer, M. | | Deposit date: | 2015-03-06 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Clickable 4-Oxo-beta-lactam-Based Selective Probing for Human Neutrophil Elastase Related Proteomes.
ChemMedChem, 11, 2016
|
|
7RC3
 
 | | Aeronamide N-methyltransferase, AerE (Y137F) | | Descriptor: | ASPARTIC ACID, CALCIUM ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Cogan, D.P, Reyes, R, Nair, S.K. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure and mechanism for iterative amide N -methylation in the biosynthesis of channel-forming peptide cytotoxins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4YN5
 
 | | Catalytic domain of Bacillus sp. JAMB-750 GH26 Endo-beta-1,4-mannanase | | Descriptor: | CACODYLATE ION, Mannan endo-1,4-beta-mannosidase | | Authors: | Shimane, Y, Ohta, Y, Usami, R, Hatada, Y. | | Deposit date: | 2015-03-09 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus sp. JAMB-750 GH26 Endo-beta-1,4-mannanase
To Be Published
|
|
6QLC
 
 | | The ssDNA-binding RNA polymerase cofactor Drc from Pseudomonas phage LUZ7 | | Descriptor: | PHOSPHATE ION, ssDNA binding RNA Polymerase cofactor | | Authors: | De Zitter, E, Boon, M, De Smet, J, Lavigne, R, Van Meervelt, L. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 'Drc', a structurally novel ssDNA-binding transcription regulator of N4-related bacterial viruses.
Nucleic Acids Res., 48, 2020
|
|
