4F9K
 
 | | Crystal Structure of human cAMP-dependent protein kinase type I-beta regulatory subunit (fragment 11-73), Northeast Structural Genomics Consortium (NESG) Target HR8613A | | Descriptor: | cAMP-dependent protein kinase type I-beta regulatory subunit | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Shastry, R, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-18 | | Release date: | 2012-06-06 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of human cAMP-dependent protein kinase type I-beta regulatory subunit (fragment 11-73), Northeast Structural Genomics Consortium (NESG) Target HR8613A
To be Published
|
|
7QNG
 
 | | Structure of a MHC I-Tapasin-ERp57 complex | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Mueller, I.K, Thomas, C, Trowitzsch, S, Tampe, R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of an MHC I-tapasin-ERp57 editing complex defines chaperone promiscuity.
Nat Commun, 13, 2022
|
|
6UUS
 
 | | CryoEM Structure of the active Adrenomedullin 2 receptor G protein complex with adrenomedullin peptide | | Descriptor: | ADM, Calcitonin gene-related peptide type 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Liang, Y.L, Sexton, P, Danev, R. | | Deposit date: | 2019-10-31 | | Release date: | 2020-04-01 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure and Dynamics of Adrenomedullin Receptors AM1and AM2Reveal Key Mechanisms in the Control of Receptor Phenotype by Receptor Activity-Modifying Proteins.
Acs Pharmacol Transl Sci, 3, 2020
|
|
6UVA
 
 | | CryoEM Structure of the active Adrenomedullin 2 receptor G protein complex with adrenomedullin 2 peptide | | Descriptor: | Calcitonin gene-related peptide type 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Belousoff, M.J, Liang, Y.L, Sexton, P, Danev, R. | | Deposit date: | 2019-11-01 | | Release date: | 2020-04-01 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structure and Dynamics of Adrenomedullin Receptors AM1and AM2Reveal Key Mechanisms in the Control of Receptor Phenotype by Receptor Activity-Modifying Proteins.
Acs Pharmacol Transl Sci, 3, 2020
|
|
6UWJ
 
 | |
6UV5
 
 | |
6UWH
 
 | |
6O29
 
 | | Crystal structure of 4493 Fab in complex with circumsporozoite protein NPDP and anti-kappa VHH domain | | Descriptor: | 1,2-ETHANEDIOL, 4493 Fab heavy chain, 4493 Kappa light chain, ... | | Authors: | Scally, S.W, Bosch, A, Prieto, K, Murugan, R, Wardemann, H, Julien, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs.
Nat. Med., 26, 2020
|
|
7Q9E
 
 | | CYP106A1 | | Descriptor: | COBALT (II) ION, Cytochrome P450, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Carius, Y, Hutter, M, Kiss, F, Bernhardt, R, Lancaster, C.R.D. | | Deposit date: | 2021-11-12 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural comparison of the cytochrome P450 enzymes CYP106A1 and CYP106A2 provides insight into their differences in steroid conversion.
Febs Lett., 596, 2022
|
|
6O2R
 
 | | Deacetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O2A
 
 | | Crystal structure of 4493 Fab in complex with circumsporozoite protein NDN and anti-kappa VHH domain | | Descriptor: | 1,2-ETHANEDIOL, 4493 Fab heavy chain, 4493 Kappa light chain, ... | | Authors: | Scally, S.W, Bosch, A, Prieto, K, Murugan, R, Wardemann, H, Julien, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs.
Nat. Med., 26, 2020
|
|
7QPD
 
 | | Structure of the human MHC I peptide-loading complex editing module | | Descriptor: | Beta-2-microglobulin, Calreticulin, HLA class I histocompatibility antigen, ... | | Authors: | Domnick, A, Susac, L, Trowitzsch, S, Thomas, C, Tampe, R. | | Deposit date: | 2022-01-03 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Molecular basis of MHC I quality control in the peptide loading complex.
Nat Commun, 13, 2022
|
|
6NR8
 
 | | hTRiC-hPFD Class6 | | Descriptor: | Prefoldin subunit 1, Prefoldin subunit 2, Prefoldin subunit 3, ... | | Authors: | Gestaut, D.R, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | Deposit date: | 2019-01-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
8ASH
 
 | | Crystal structure of d(CCGGGGTACCCCGG) with XRB | | Descriptor: | 4-[(~{E})-(3,6-dimethyl-1,3-benzothiazol-2-yl)iminomethyl]-~{N},~{N}-dimethyl-aniline, DNA (5'-D(*CP*CP*GP*GP*GP*GP*TP*AP*CP*CP*CP*CP*GP*G)-3') | | Authors: | Sbirkova-Dimitrova, H.I, Shivachev, B.L, Rusev, R, Kuvandjiev, N, Heroux, A, Doukov, T. | | Deposit date: | 2022-08-19 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.837 Å) | | Cite: | Structural Characterization of Alzheimer DNA Promoter Sequences from the Amyloid Precursor Gene in the Presence of Thioflavin T and Analogs
Crystals, 12, 2022
|
|
6NYW
 
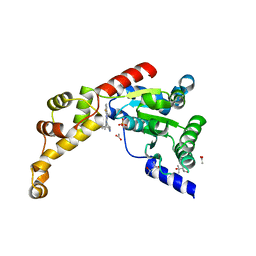 | | Structure of spastin AAA domain N527C mutant in complex with 8-fluoroquinazoline-based inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, N-(5-tert-butyl-1H-pyrazol-3-yl)-8-fluoro-2-[(2R)-2-methylpiperazin-1-yl]quinazolin-4-amine, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-02-12 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Designing Allele-Specific Inhibitors of Spastin, a Microtubule-Severing AAA Protein.
J. Am. Chem. Soc., 141, 2019
|
|
8ASK
 
 | | Crystal structure of d(GCCCACCACGGC) | | Descriptor: | DNA (5'-D(P*GP*CP*CP*CP*AP*CP*CP*AP*CP*GP*GP*C)-3'), DNA (5'-D(P*GP*CP*CP*GP*TP*GP*GP*TP*GP*GP*GP*C)-3') | | Authors: | Sbirkova-Dimitrova, H.I, Shivachev, B.L, Rusev, R, Heroux, A, Doukov, T, Kuvandjiev, N. | | Deposit date: | 2022-08-19 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | Structural Characterization of Alzheimer DNA Promoter Sequences from the Amyloid Precursor Gene in the Presence of Thioflavin T and Analogs
Crystals, 12, 2022
|
|
6O0Y
 
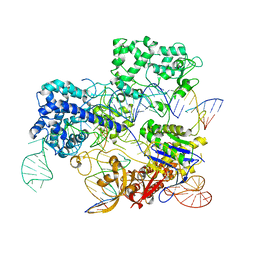 | | Conformational states of Cas9-sgRNA-DNA ternary complex in the presence of magnesium | | Descriptor: | 3' product of target strand DNA, 5' product of target strand DNA, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Zhu, X, Clarke, R, Puppala, A.K, Chittori, S, Merk, A, Merrill, B.J, Simonovic, M, Subramaniam, S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6O25
 
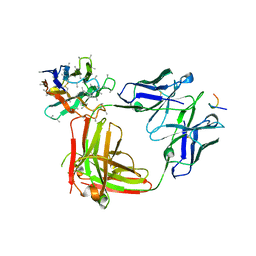 | | Crystal structure of 3945 Fab in complex with circumsporozoite protein NANP3 and anti-Kappa VHH domain | | Descriptor: | 3945 Fab heavy chain, 3945 Kappa light chain, Anti-Kappa VHH domain, ... | | Authors: | Scally, S.W, Bosch, A, Prieto, K, Murugan, R, Wardemann, H, Julien, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs.
Nat. Med., 26, 2020
|
|
6UQG
 
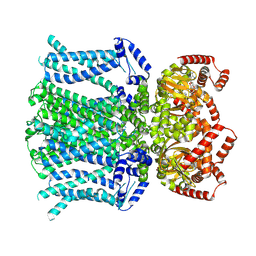 | | Human HCN1 channel Y289D mutant | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Lee, C.-H, MacKinnon, R. | | Deposit date: | 2019-10-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Voltage Sensor Movements during Hyperpolarization in the HCN Channel.
Cell, 179, 2019
|
|
6UR0
 
 | | Crystal structure of ChoE D285N mutant acyl-enzyme | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, ChoE, GLYCEROL | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2019-10-21 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the putative bacterial acetylcholinesterase ChoE and its substrate inhibition mechanism.
J.Biol.Chem., 295, 2020
|
|
7QVP
 
 | | Human collided disome (di-ribosome) stalled on XBP1 mRNA | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Denk, T.G, Tesina, P, Beckmann, R. | | Deposit date: | 2022-01-22 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A distinct mammalian disome collision interface harbors K63-linked polyubiquitination of uS10 to trigger hRQT-mediated subunit dissociation.
Nat Commun, 13, 2022
|
|
6OXD
 
 | | Structure of Mycobacterium tuberculosis methylmalonyl-CoA mutase with adenosyl cobalamin | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Itaconyl coenzyme A, ... | | Authors: | Purchal, M, Ruetz, M, Banerjee, R, Koutmos, M. | | Deposit date: | 2019-05-13 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Itaconyl-CoA forms a stable biradical in methylmalonyl-CoA mutase and derails its activity and repair.
Science, 366, 2019
|
|
6OY1
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-26 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[({4-[(1R)-1,2-dihydroxyethyl]phenyl}sulfonyl)(2-ethylbutyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
4FI4
 
 | | Crystal structure of mannonate dehydratase PRK15072 (TARGET EFI-502214) from Caulobacter sp. K31 | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-06-07 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Enolase Prk15072 (Target Efi-502214) from Caulobacter Sp. K31
To be Published
|
|
6UUW
 
 | | Structure of human ATP citrate lyase E599Q mutant in complex with Mg2+, citrate, ATP and CoA | | Descriptor: | (2S)-2-hydroxy-2-[2-oxo-2-(phosphonooxy)ethyl]butanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2019-11-01 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Molecular basis for acetyl-CoA production by ATP-citrate lyase
Nat.Struct.Mol.Biol., 27, 2020
|
|
