4BAD
 
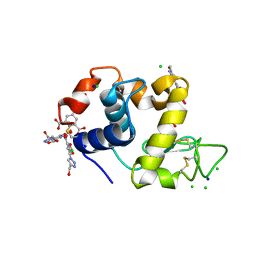 | | Hen egg-white lysozyme structure in complex with the europium tris- hydroxymethyltriazoledipicolinate complex at 1.35 A resolution. | | Descriptor: | 4-(4-(hydroxymethyl)-1h-1,2,3-triazol-1-yl)pyridine-2,6-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Talon, R, Kahn, R, Gautier, A, Nauton, L, Girard, E. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-14 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Clicked Europium Dipicolinate Complexes for Protein X-Ray Structure Determination.
Chem.Commun.(Camb.), 48, 2012
|
|
4JXI
 
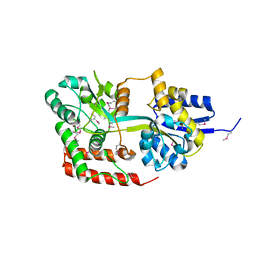 | | Directed evolution and rational design of a de novo designed esterase toward improved catalysis. Northeast Structural Genomics Consortium (NESG) Target OR184 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, BROMIDE ION, GLYCEROL, ... | | Authors: | Kuzin, A, Smith, M.D, Richter, F, Lew, S, Seetharaman, R, Bryan, C, Lech, Z, Kiss, G, Moretti, R, Maglaqui, M, Xiao, R, Kohan, E, Smith, M, Everett, J.K, Nguyen, R, Pande, V, Hilvert, D, Kornhaber, G, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Directed evolution and rational design of a de novo designed esterase toward improved catalysis. Northeast Structural Genomics Consortium (NESG) Target OR184
To be Published
|
|
2F1K
 
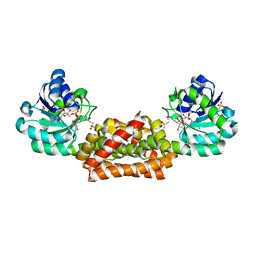 | | Crystal structure of Synechocystis arogenate dehydrogenase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, prephenate dehydrogenase | | Authors: | Legrand, P, Dumas, R, Seux, M, Rippert, P, Ravelli, R, Ferrer, J.-L, Matringe, M. | | Deposit date: | 2005-11-14 | | Release date: | 2006-05-09 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biochemical Characterization and Crystal Structure of Synechocystis Arogenate Dehydrogenase Provide Insights into Catalytic Reaction
Structure, 14, 2006
|
|
1IED
 
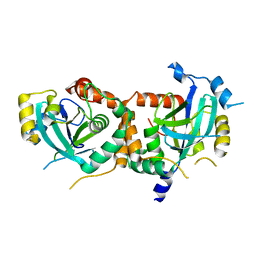 | | CRYSTAL STRUCTURE OF THE CATALYTIC SITE MUTANT (H157E) OF THE HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | CAPSID PROTEIN P40: ASSEMBLIN PROTEASE | | Authors: | Khayat, R, Batra, R, Massariol, M.J, Lagace, L, Tong, L. | | Deposit date: | 2001-04-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigating the role of histidine 157 in the catalytic activity of human cytomegalovirus protease.
Biochemistry, 40, 2001
|
|
1E0V
 
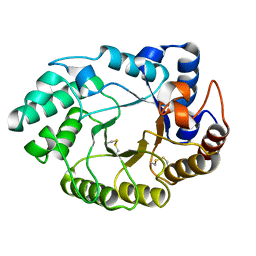 | | Xylanase 10A from Sreptomyces lividans. cellobiosyl-enzyme intermediate at 1.7 A | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
4BAR
 
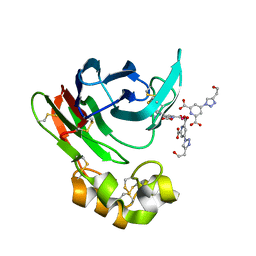 | | Thaumatin from Thaumatococcus daniellii structure in complex with the europium tris-hydroxyethyltriazoledipicolinate complex at 1.20 A resolution. | | Descriptor: | 4-(4-(2-hydroxyethyl)-1H-1,2,3-triazol-1-yl)pyridine-2,6-dicarboxylic acid, EUROPIUM (III) ION, THAUMATIN-1 | | Authors: | Talon, R, Kahn, R, Gautier, A, Nauton, L, Girard, E. | | Deposit date: | 2012-09-14 | | Release date: | 2012-11-14 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Clicked europium dipicolinate complexes for protein X-ray structure determination.
Chem. Commun. (Camb.), 48, 2012
|
|
1M4Y
 
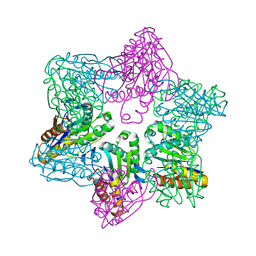 | | Crystal structure of HslV from Thermotoga maritima | | Descriptor: | ATP-dependent protease hslV, SODIUM ION | | Authors: | Song, H.K, Ramachandran, R, Bochtler, M.B, Hartmann, C, Azim, M.K, Huber, R. | | Deposit date: | 2002-07-05 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Isolation and characterization of the prokaryotic proteasome homolog HslVU (ClpQY) from Thermotoga maritima and the crystal structure of HslV.
BIOPHYS.CHEM., 100, 2003
|
|
1MKJ
 
 | | Human Kinesin Motor Domain With Docked Neck Linker | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN HEAVY CHAIN, MAGNESIUM ION, ... | | Authors: | Sindelar, C.V, Budny, M.J, Rice, S, Naber, N, Fletterick, R, Cooke, R. | | Deposit date: | 2002-08-29 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two conformations in the human kinesin power stroke defined by X-ray crystallography and EPR spectroscopy.
Nat.Struct.Biol., 9, 2002
|
|
1QMG
 
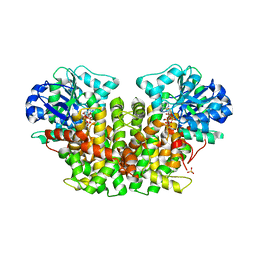 | | Acetohydroxyacid isomeroreductase complexed with its reaction product dihydroxy-methylvalerate, manganese and ADP-ribose. | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHORIBOSE, 2,3-DIHYDROXY-VALERIANIC ACID, ACETOHYDROXY-ACID ISOMEROREDUCTASE, ... | | Authors: | Thomazeau, K, Dumas, R, Halgand, F, Douce, R, Biou, V. | | Deposit date: | 1999-09-28 | | Release date: | 2000-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Spinach Acetohydroxyacid Isomeroreductase Complexed with its Product of Reaction Dihydroxy-Methylvalerate, Manganese and Adp-Ribose
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2FMN
 
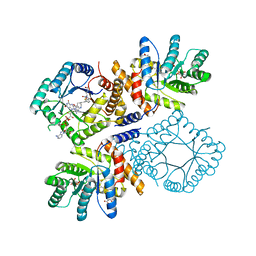 | | Ala177Val mutant of E. coli Methylenetetrahydrofolate Reductase complex with LY309887 | | Descriptor: | 5,10-methylenetetrahydrofolate reductase, FLAVIN-ADENINE DINUCLEOTIDE, N-[(5-{2-[(6R)-2-AMINO-4-OXO-3,4,5,6,7,8-HEXAHYDROPYRIDO[2,3-D]PYRIMIDIN-6-YL]ETHYL}-2-THIENYL)CARBONYL]-L-GLUTAMIC ACID | | Authors: | Pejchal, R, Campbell, E, Guenther, B.D, Lennon, B.W, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Perturbations in the Ala -> Val Polymorphism of Methylenetetrahydrofolate Reductase: How Binding of Folates May Protect against Inactivation
Biochemistry, 45, 2006
|
|
4BAF
 
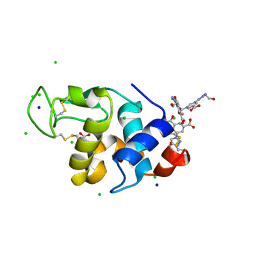 | | Hen egg-white lysozyme structure in complex with the europium tris- hydroxyethyltriazoledipicolinate complex at 1.51 A resolution. | | Descriptor: | 4-(4-(2-hydroxyethyl)-1H-1,2,3-triazol-1-yl)pyridine-2,6-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Talon, R, Kahn, R, Gautier, A, Nauton, L, Girard, E. | | Deposit date: | 2012-09-14 | | Release date: | 2012-11-14 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.507 Å) | | Cite: | Clicked Europium Dipicolinate Complexes for Protein X-Ray Structure Determination.
Chem.Commun.(Camb.), 48, 2012
|
|
2FPN
 
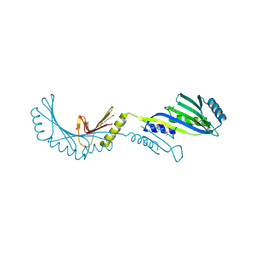 | |
2FMO
 
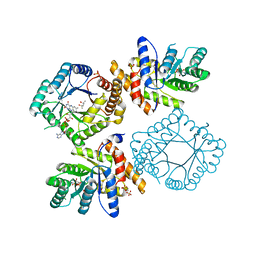 | | Ala177Val mutant of E. coli Methylenetetrahydrofolate Reductase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5,10-methylenetetrahydrofolate reductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Pejchal, R, Campbell, E, Guenther, B.D, Lennon, B.W, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Perturbations in the Ala -> Val Polymorphism of Methylenetetrahydrofolate Reductase: How Binding of Folates May Protect against Inactivation
Biochemistry, 45, 2006
|
|
4BAP
 
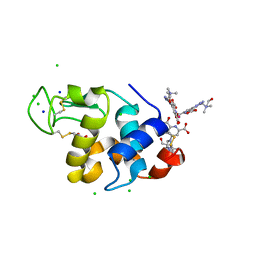 | | Hen egg-white lysozyme structure in complex with the europium tris- hydroxyethylcholinetriazoledipicolinate complex at 1.21 A resolution. | | Descriptor: | ACETATE ION, CHLORIDE ION, EUROPIUM (III) ION, ... | | Authors: | Talon, R, Kahn, R, Gautier, A, Nauton, L, Girard, E. | | Deposit date: | 2012-09-14 | | Release date: | 2012-11-14 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.207 Å) | | Cite: | Clicked Europium Dipicolinate Complexes for Protein X-Ray Structure Determination.
Chem.Commun.(Camb.), 48, 2012
|
|
2FQ4
 
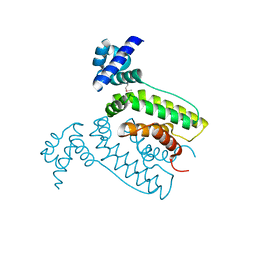 | | The crystal structure of the transcriptional regulator (TetR family) from Bacillus cereus | | Descriptor: | Transcriptional regulator, TetR family | | Authors: | Zhang, R, Wu, R, Moy, S, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-17 | | Release date: | 2006-02-28 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The crystal structure of the transcriptional regulator (TetR family) from Bacillus cereus
To be Published
|
|
2FQ3
 
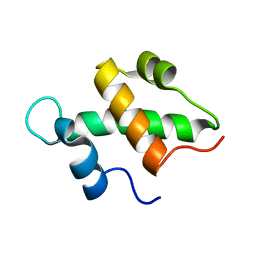 | | Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes | | Descriptor: | Transcription regulatory protein SWI3 | | Authors: | Da, G, Lenkart, J, Zhao, K, Shiekhattar, R, Cairns, B.R, Marmorstein, R. | | Deposit date: | 2006-01-17 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4CCJ
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66) in apo form | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66 | | Authors: | Chowdhury, R, Ge, W, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CCM
 
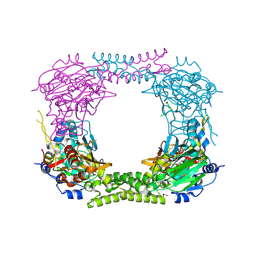 | | 60S ribosomal protein L8 histidine hydroxylase (NO66) in complex with Mn(II), N-oxalylglycine (NOG) and 60S ribosomal protein L8 (RPL8 G220C) peptide fragment (complex-1) | | Descriptor: | 1,2-ETHANEDIOL, 60S RIBOSOMAL PROTEIN L8, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, ... | | Authors: | Chowdhury, R, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
2R9M
 
 | | Cathepsin S complexed with Compound 15 | | Descriptor: | Cathepsin S, N-[(1S)-2-[(4-cyano-1-methylpiperidin-4-yl)amino]-1-(cyclohexylmethyl)-2-oxoethyl]morpholine-4-carboxamide | | Authors: | Ward, Y.D, Emmanuel, M.J, Thomson, D.S, Liu, W, Bekkali, Y, Frye, L.L, Girardot, M, Morwick, T, Young, E.R.R, Zindell, R, Hrapchak, M, DeTuri, M, White, A, Crane, K.M, White, D.M, Wang, Y, Hao, M.-H, Grygon, C.A, Labadia, M.E, Wildeson, J, Freeman, D, Nelson, R, Capolino, A, Peterson, J.D, Raymond, E.L, Brown, M.L, Spero, D.M. | | Deposit date: | 2007-09-13 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Design and Synthesis of Reversible Inhibitors of Cathepsin S: alpha,alpha-Disubstitution at the P1 Residue Provides Potent Inhibitors in Cellular Assays and In Vivo Models of Antigen Presentation
To be Published
|
|
6XGR
 
 | | YSD1 major tail protein | | Descriptor: | YSD1_22 major tail protein | | Authors: | Hardy, J.M, Dunstan, R, Venugopal, H, Lithgow, T.J, Coulibaly, F.J. | | Deposit date: | 2020-06-17 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The architecture and stabilisation of flagellotropic tailed bacteriophages.
Nat Commun, 11, 2020
|
|
7D3I
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with MI-23 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[3-[3,5-bis(fluoranyl)phenyl]propanoyl]-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase | | Authors: | Zeng, R, Li, Y.S, Qiao, J.X, Wang, Y.F, Yang, S.Y, Lei, J. | | Deposit date: | 2020-09-19 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
4CCO
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66 S373C) in complex with Mn(II), N-oxalylglycine (NOG) and 60S ribosomal protein L8 (RPL8 G214C) peptide fragment (complex-3) | | Descriptor: | 1,2-ETHANEDIOL, 60S RIBOSOMAL PROTEIN L8, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, ... | | Authors: | Chowdhury, R, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
2R71
 
 | | Crystal structure of the complex of bovine C-lobe with inositol at 2.1A resolution | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Mir, R, Jain, R, Kumar, S, Sinha, M, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-09-07 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of the complex of bovine C-lobe with inositol at 2.1A resolution
To be Published
|
|
2R9N
 
 | | Cathepsin S complexed with Compound 26 | | Descriptor: | Cathepsin S, N-[(1S)-2-{[(3S)-1-benzyl-3-cyanopyrrolidin-3-yl]amino}-1-(cyclohexylmethyl)-2-oxoethyl]morpholine-4-carboxamide | | Authors: | Ward, Y.D, Emmanuel, M.J, Thomson, D.S, Liu, W, Bekkali, Y, Frye, L.L, Girardot, M, Morwick, T, Young, E.R.R, Zindell, R, Hrapchak, M, DeTuri, M, White, A, Crane, K.M, White, D.M, Wang, Y, Hao, M.-H, Grygon, C.A, Labadia, M.E, Wildeson, J, Freeman, D, Nelson, R, Capolino, A, Peterson, J.D, Raymond, E.L, Brown, M.L, Spero, D.M. | | Deposit date: | 2007-09-13 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Reversible Inhibitors of Cathepsin S: alpha,alpha-Disubstitution at the P1 Residue Provides Potent Inhibitors in Cellular Assays and In Vivo Models of Antigen Presentation
to be published
|
|
2R9O
 
 | | Cathepsin S complexed with Compound 8 | | Descriptor: | Cathepsin S, N-[(1S)-2-{[(1R)-2-(benzyloxy)-1-cyano-1-methylethyl]amino}-1-(cyclohexylmethyl)-2-oxoethyl]morpholine-4-carboxamide | | Authors: | Ward, Y.D, Emmanuel, M.J, Thomson, D.S, Liu, W, Bekkali, Y, Frye, L.L, Girardot, M, Morwick, T, Young, E.R.R, Zindell, R, Hrapchak, M, DeTuri, M, White, A, Crane, K.M, White, D.M, Wang, Y, Hao, M.-H, Grygon, C.A, Labadia, M.E, Wildeson, J, Freeman, D, Nelson, R, Capolino, A, Peterson, J.D, Raymond, E.L, Brown, M.L, Spero, D.M. | | Deposit date: | 2007-09-13 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Reversible Inhibitors of Cathepsin S: alpha,alpha-Disubstitution at the P1 Residue Provides Potent Inhibitors in Cellular Assays and In Vivo Models of Antigen Presentation
to be published
|
|
