8EXK
 
 | |
8TU1
 
 | |
8F3Q
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) Y460A variant apo form from Enterococcus faecium | | 分子名称: | Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Schoenle, M.V, Choy, M.S, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
1HH5
 
 | | cytochrome c7 from Desulfuromonas acetoxidans | | 分子名称: | CYTOCHROME C7, HEME C | | 著者 | Czjzek, M, Haser, R, Arnoux, P, Shepard, W. | | 登録日 | 2000-12-21 | | 公開日 | 2001-05-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of Cytochrome C7 from Desulfuromonas Acetoxidans at 1.9A Resolutio N
Acta Crystallogr.,Sect.D, 57, 2001
|
|
8TU0
 
 | |
6GFA
 
 | | Structure of Nucleotide binding domain of HSP110, ATP and Mg2+ complexed | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 105 kDa, MAGNESIUM ION | | 著者 | Gonzalez, D, Gotthard, G, Gozzi, G.J, Seigneuric, R, Neiers, F, Briand, L, Garrido, C. | | 登録日 | 2018-04-29 | | 公開日 | 2019-05-08 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Selecting the first chemical molecule inhibitor of HSP110 for colorectal cancer therapy.
Cell Death Differ., 27, 2020
|
|
6QZQ
 
 | |
4ZOB
 
 | | Crystal Structure of beta-glucosidase from Listeria innocua in complex with gluconolactone | | 分子名称: | D-glucono-1,5-lactone, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | 登録日 | 2015-05-06 | | 公開日 | 2016-05-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
6QMA
 
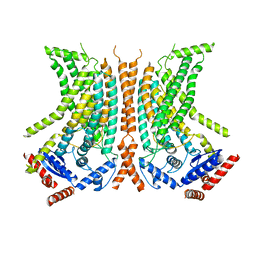 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in nanodisc (intermediate state) | | 分子名称: | CALCIUM ION, Predicted protein | | 著者 | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | 登録日 | 2019-02-01 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
3ZE7
 
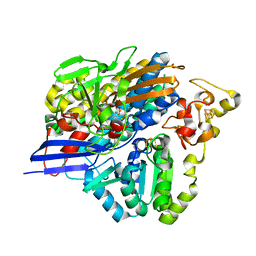 | | 3D structure of the Ni-Fe-Se hydrogenase from D. vulgaris Hildenborough in the reduced state at 1.95 Angstroms | | 分子名称: | CARBONMONOXIDE-(DICYANO) IRON, FE (II) ION, HYDROSULFURIC ACID, ... | | 著者 | Marques, M.C, Coelho, R, Pereira, I.A.C, Matias, P.M. | | 登録日 | 2012-12-03 | | 公開日 | 2013-06-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Redox State-Dependent Changes in the Crystal Structure of [Nifese] Hydrogenase from Desulfovibrio Vulgaris Hildenborough
Int.J.Hydrogen Energy, 38, 2013
|
|
8U1J
 
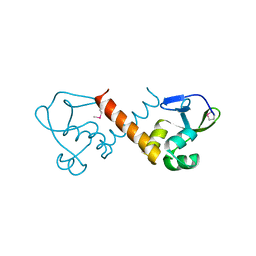 | |
1H68
 
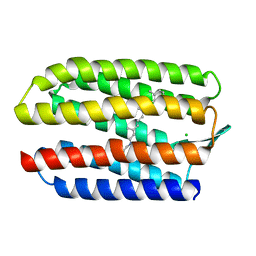 | | sensory rhodopsin II | | 分子名称: | CHLORIDE ION, RETINAL, SENSORY RHODOPSIN II | | 著者 | Royant, A, Nollert, P, Edman, K, Neutze, R, Landau, E.M, Pebay-Peyroula, E, Navarro, J. | | 登録日 | 2001-06-08 | | 公開日 | 2001-08-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | X-Ray Structure of Sensory Rhodopsin II at 2.1 A Resolution
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4WYG
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis complexed with a fragment hit | | 分子名称: | 1,2-ETHANEDIOL, 1-{4-[(4-chloro-1H-pyrazol-1-yl)methyl]phenyl}methanamine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Dai, R, Finzel, B.C. | | 登録日 | 2014-11-17 | | 公開日 | 2015-07-08 | | 最終更新日 | 2015-07-22 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Fragment-Based Exploration of Binding Site Flexibility in Mycobacterium tuberculosis BioA.
J.Med.Chem., 58, 2015
|
|
4WUR
 
 | |
6QP6
 
 | | Cryo-EM structure of calcium-bound mTMEM16F lipid scramblase in digitonin | | 分子名称: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6, CALCIUM ION | | 著者 | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | 登録日 | 2019-02-13 | | 公開日 | 2019-03-06 | | 最終更新日 | 2019-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
4WWH
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM MYCOBACTERIUM SMEGMATIS (MSMEG_1704, TARGET EFI-510967) WITH BOUND D-GALACTOSE | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ABC transporter, beta-D-galactopyranose | | 著者 | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-11-11 | | 公開日 | 2014-11-26 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM MYCOBACTERIUM SMEGMATIS (MSMEG_1704, TARGET EFI-510967) WITH BOUND D-GALACTOSE
To be published
|
|
4JZF
 
 | | Structure of factor VIIA in complex with the inhibitor 2-{2-[(3-carbamoylphenyl)carbamoyl]-6-methoxypyridin-3-yl}-5-{[(2S)-1-hydroxy-3,3-dimethylbutan-2-yl]carbamoyl}benzoic acid | | 分子名称: | 2-{2-[(3-carbamoylphenyl)carbamoyl]-6-methoxypyridin-3-yl}-5-{[(2S)-1-hydroxy-3,3-dimethylbutan-2-yl]carbamoyl}benzoic acid, CALCIUM ION, Factor VIIa (Heavy Chain), ... | | 著者 | Wei, A, Anumula, R. | | 登録日 | 2013-04-02 | | 公開日 | 2013-08-21 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Discovery of nonbenzamidine factor VIIa inhibitors using a biaryl acid scaffold.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3SM3
 
 | | Crystal Structure of SAM-dependent methyltransferases Q8PUK2_METMA from Methanosarcina mazei. Northeast Structural Genomics Consortium Target MaR262. | | 分子名称: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, SAM-dependent methyltransferases | | 著者 | Vorobiev, S, Neely, H, Seetharaman, J, Snyder, A, Patel, P, Xiao, R, Ciccosanti, C, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-06-27 | | 公開日 | 2011-07-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.197 Å) | | 主引用文献 | Crystal Structure of SAM-dependent methyltransferases Q8PUK2_METMA from Methanosarcina mazei.
To be Published, 2009
|
|
1GTQ
 
 | | 6-PYRUVOYL TETRAHYDROPTERIN SYNTHASE | | 分子名称: | 6-PYRUVOYL TETRAHYDROPTERIN SYNTHASE, ZINC ION | | 著者 | Nar, H, Huber, R, Heizmann, C.W, Thoeny, B, Buergisser, D. | | 登録日 | 1995-09-16 | | 公開日 | 1996-04-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Three-dimensional structure of 6-pyruvoyl tetrahydropterin synthase, an enzyme involved in tetrahydrobiopterin biosynthesis.
EMBO J., 13, 1994
|
|
1GRL
 
 | | THE CRYSTAL STRUCTURE OF THE BACTERIAL CHAPERONIN GROEL AT 2.8 ANGSTROMS | | 分子名称: | GROEL (HSP60 CLASS) | | 著者 | Braig, K, Otwinowski, Z, Hegde, R, Boisvert, D.C, Joachimiak, A, Horwich, A.L, Sigler, P.B. | | 登録日 | 1995-03-07 | | 公開日 | 1995-10-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The crystal structure of the bacterial chaperonin GroEL at 2.8 A.
Nature, 371, 1994
|
|
8EP1
 
 | |
8EOW
 
 | |
4X2D
 
 | |
1GTG
 
 | | Crystal structure of the thermostable serine-carboxyl type proteinase, kumamolysin (kscp) | | 分子名称: | CALCIUM ION, KUMAMOLYSIN | | 著者 | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | 登録日 | 2002-01-15 | | 公開日 | 2002-06-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
4X3E
 
 | |
