8CN2
 
 | |
5GQV
 
 | |
5GR4
 
 | |
8C64
 
 | | Crystal structure of arsenoplatin-1/B-DNA adduct obtained upon 48 h of soaking | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PLATINUM (II) ION, arsenoplatin-1 | | 著者 | Troisi, R, Tito, G, Ferraro, G, Sica, F, Merlino, A. | | 登録日 | 2023-01-11 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | On the mechanism of action of arsenoplatins: arsenoplatin-1 binding to a B-DNA dodecamer.
Dalton Trans, 53, 2024
|
|
8C9H
 
 | | AQP7_inhibitor | | 分子名称: | Aquaporin-7, ethyl 4-[(4-pyrazol-1-ylphenyl)methylcarbamoylamino]benzoate | | 著者 | Huang, P, Venskutonyte, R, Gourdon, P, Lindkvist-Petersson, K. | | 登録日 | 2023-01-22 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Molecular basis for human aquaporin inhibition.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8C62
 
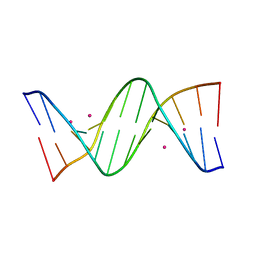 | | Crystal structure of cisplatin/B-DNA adduct | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PLATINUM (II) ION | | 著者 | Troisi, R, Tito, G, Ferraro, G, Sica, F, Merlino, A. | | 登録日 | 2023-01-11 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | On the mechanism of action of arsenoplatins: arsenoplatin-1 binding to a B-DNA dodecamer.
Dalton Trans, 53, 2024
|
|
1IMF
 
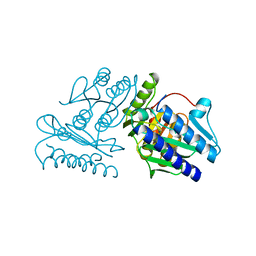 | |
2YMD
 
 | | Crystal structure of a mutant binding protein (5HTBP-AChBP) in complex with serotonin (5-hydroxytryptamine) | | 分子名称: | GLYCEROL, PHOSPHATE ION, SEROTONIN, ... | | 著者 | Kesters, D, Thompson, A.J, Brams, M, Elk, R.v, Spurny, R, Geitmann, M, Villalgordo, J.M, Guskov, A, Danielson, U.H, Lummis, S.C.R, Smit, A.B, Ulens, C. | | 登録日 | 2012-10-09 | | 公開日 | 2012-12-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structural Basis of Ligand Recognition in 5-Ht(3) Receptors.
Embo Rep., 14, 2013
|
|
8C9L
 
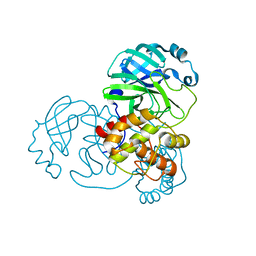 | |
8C9O
 
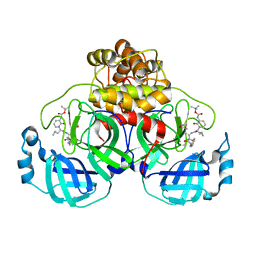 | |
1N62
 
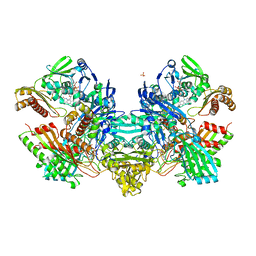 | | Crystal Structure of the Mo,Cu-CO Dehydrogenase (CODH), n-butylisocyanide-bound state | | 分子名称: | CU(I)-S-MO(IV)(=O)O-NBIC CLUSTER, Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, ... | | 著者 | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | 登録日 | 2002-11-08 | | 公開日 | 2002-12-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4MIV
 
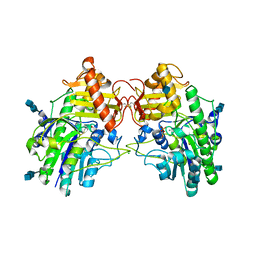 | | Crystal Structure of Sulfamidase, Crystal Form L | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Sidhu, N.S, Uson, I, Schreiber, K, Proepper, K, Becker, S, Sheldrick, G.M, Gaertner, J, Kraetzner, R, Steinfeld, R. | | 登録日 | 2013-09-02 | | 公開日 | 2014-05-14 | | 最終更新日 | 2021-06-02 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of sulfamidase provides insight into the molecular pathology of mucopolysaccharidosis IIIA.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1N60
 
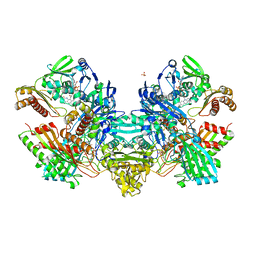 | | Crystal Structure of the Cu,Mo-CO Dehydrogenase (CODH); Cyanide-inactivated Form | | 分子名称: | Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, Carbon monoxide dehydrogenase small chain, ... | | 著者 | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | 登録日 | 2002-11-08 | | 公開日 | 2002-12-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.19 Å) | | 主引用文献 | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5GQY
 
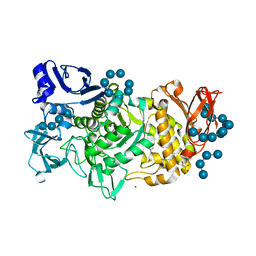 | |
8COD
 
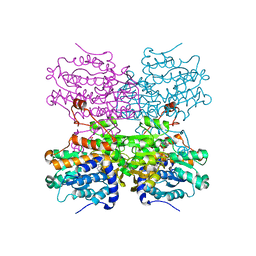 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Mus musculus in complex with inosine | | 分子名称: | Adenosylhomocysteinase, INOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Saleem-Batcha, R, Popadic, D, Koeppl, L.H, Andexer, J.N. | | 登録日 | 2023-02-27 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-09-18 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Structure, function and substrate preferences of archaeal S-adenosyl-L-homocysteine hydrolases.
Commun Biol, 7, 2024
|
|
8CKH
 
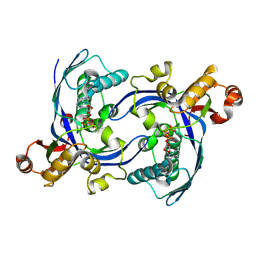 | |
1IMC
 
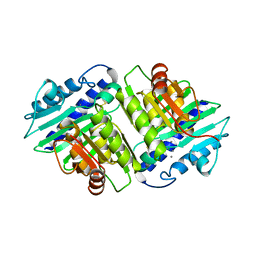 | |
8F41
 
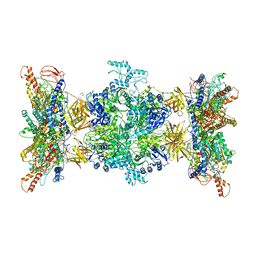 | | 3-methylcrotonyl-CoA carboxylase in filament, alpha-subunit centered | | 分子名称: | 3-methylcrotonyl-CoA carboxylase, alpha-subunit, beta-subunit, ... | | 著者 | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | 登録日 | 2022-11-10 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
8CMV
 
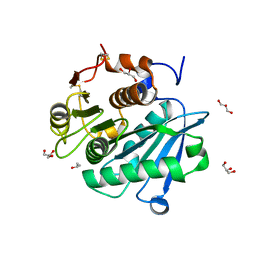 | |
2ZCE
 
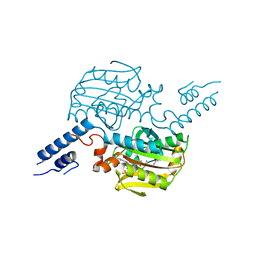 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with pyrrolysine and an ATP analogue | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PYRROLYSINE, ... | | 著者 | Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-11-08 | | 公開日 | 2008-04-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystallographic Studies on Multiple Conformational States of Active-site Loops in Pyrrolysyl-tRNA Synthetase
J.Mol.Biol., 378, 2008
|
|
2ZIN
 
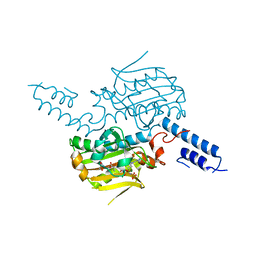 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with BocLys and an ATP analogue | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, N~6~-(tert-butoxycarbonyl)-L-lysine, ... | | 著者 | Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2008-02-19 | | 公開日 | 2008-12-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Multistep Engineering of Pyrrolysyl-tRNA Synthetase to Genetically Encode N(varepsilon)-(o-Azidobenzyloxycarbonyl) lysine for Site-Specific Protein Modification
Chem.Biol., 15, 2008
|
|
4MP4
 
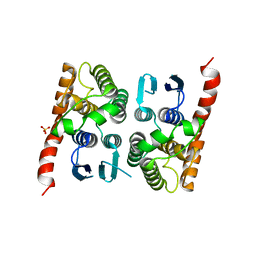 | | Crystal structure of a glutathione transferase family member from Acinetobacter baumannii, Target EFI-501785, apo structure | | 分子名称: | Glutathione S-transferase, SULFATE ION | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-09-12 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.498 Å) | | 主引用文献 | Crystal structure of a glutathione transferase family member from Acinetobacter baumannii, Target EFI-501785, apo structure
To be Published
|
|
6Y0P
 
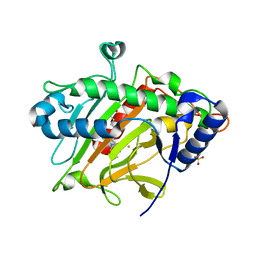 | | isopenicillin N synthase in complex with IPN and Fe using FT-SSX methods | | 分子名称: | FE (III) ION, ISOPENICILLIN N, Isopenicillin N synthase, ... | | 著者 | Rabe, P, Beale, J.H, Lang, P.A, Dirr, A.S, Leissing, T.M, Butryn, A, Aller, P, Kamps, J.J.A.G, Axford, D, McDonough, M.A, Orville, A.M, Owen, R, Schofield, C.J. | | 登録日 | 2020-02-10 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | X-ray free-electron laser studies reveal correlated motion during isopenicillin N synthase catalysis.
Sci Adv, 7, 2021
|
|
6PTO
 
 | | Structure of Ctf4 trimer in complex with three CMG helicases | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA polymerase alpha-binding protein, ... | | 著者 | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | 登録日 | 2019-07-16 | | 公開日 | 2019-11-20 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
1HM5
 
 | |
