1M71
 
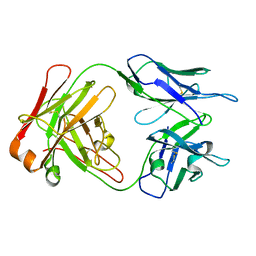 | | Crystal structure of a Monoclonal Fab Specific for Shigella Flexneri Y lipopolysaccharide | | Descriptor: | heavy chain of the monoclonal antibody Fab SYA/J6, light chain of the monoclonal antibody Fab SYA/J6 | | Authors: | Vyas, N.K, Vyas, M.N, Chervenak, M.C, Johnson, M.A, Pinto, B.M, Bundle, D.R, Quiocho, F.A. | | Deposit date: | 2002-07-18 | | Release date: | 2003-07-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Recognition of Oligosaccharide Epitopes by a Monoclonal Fab Specific for
Shigella flexneri Y Lipopolysaccharide: X-ray Structures and Thermodyanamics
Biochemistry, 41, 2002
|
|
1M7D
 
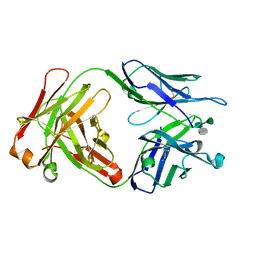 | | Crystal structure of a Monoclonal Fab Specific for Shigella flexneri Y Lipopolysaccharide complexed with a trisaccharide | | Descriptor: | alpha-L-rhamnopyranose-(1-3)-alpha-L-Olivopyranose-(1-3)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, heavy chain of the monoclonal antibody Fab SYA/J6, light chain of the monoclonal antibody Fab SYA/J6 | | Authors: | Vyas, N.K, Vyas, M.N, Chervenak, M.C, Johnson, M.A, Pinto, B.M, Bundle, D.R, Quiocho, F.A. | | Deposit date: | 2002-07-19 | | Release date: | 2003-07-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Recognition of Oligosaccharide Epitopes by a Monoclonal Fab Specific for Shigella flexneri
Y Lipopolysaccharide: X-ray Structures and Thermodynamics
Biochemistry, 41, 2002
|
|
1M7I
 
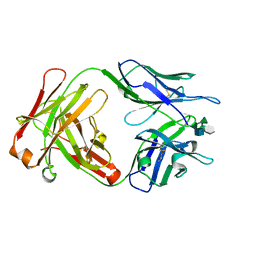 | | CRYSTAL STRUCTURE OF A MONOCLONAL FAB SPECIFIC FOR SHIGELLA FLEXNERI Y LIPOPOLYSACCHARIDE COMPLEXED WITH A PENTASACCHARIDE | | Descriptor: | alpha-L-rhamnopyranose-(1-2)-alpha-L-rhamnopyranose-(1-3)-alpha-L-rhamnopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-methyl 6-deoxy-alpha-L-rhamnopyranoside, heavy chain of the monoclonal antibody Fab SYA/J6, light chain of the monoclonal antibody Fab SYA/J6 | | Authors: | Vyas, N.K, Vyas, M.N, Chervenak, M.C, Johnson, M.A, Pinto, B.M, Bundle, D.R, Quiocho, F.A. | | Deposit date: | 2002-07-19 | | Release date: | 2003-07-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Recognition of Oligosaccharide Epitopes by a Monoclonal Fab Specific for
Shigella flexneri Y Lipopolysaccharide: X-ray Structures and Thermodynamics
Biochemistry, 41, 2002
|
|
1MAR
 
 | |
1LWS
 
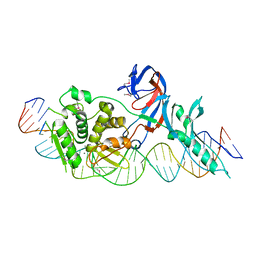 | | Crystal structure of the intein homing endonuclease PI-SceI bound to its recognition sequence | | Descriptor: | CALCIUM ION, ENDONUCLEASE PI-SCEI, PI-SceI DNA recognition region bottom strand, ... | | Authors: | Moure, C.M, Gimble, F.S, Quiocho, F.A. | | Deposit date: | 2002-06-03 | | Release date: | 2002-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the intein homing endonuclease PI-SceI bound to its recognition sequence.
Nat.Struct.Biol., 9, 2002
|
|
1EF0
 
 | |
1U5O
 
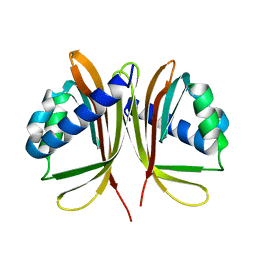 | | Structure of the D23A mutant of the nuclear transport carrier NTF2 | | Descriptor: | Nuclear transport factor 2 | | Authors: | Cushman, I, Bowman, B.R, Sowa, M.E, Lichtarge, O, Quiocho, F.A, Moore, M.S. | | Deposit date: | 2004-07-28 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Computational and biochemical identification of a nuclear pore complex binding site on the nuclear transport carrier NTF2.
J.Mol.Biol., 344, 2004
|
|
1VPT
 
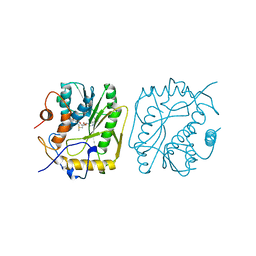 | | AS11 VARIANT OF VACCINIA VIRUS PROTEIN VP39 IN COMPLEX WITH S-ADENOSYL-L-METHIONINE | | Descriptor: | S-ADENOSYLMETHIONINE, VP39 | | Authors: | Hodel, A.E, Gershon, P.D, Shi, X, Quiocho, F.A. | | Deposit date: | 1996-03-20 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.85 A structure of vaccinia protein VP39: a bifunctional enzyme that participates in the modification of both mRNA ends.
Cell(Cambridge,Mass.), 85, 1996
|
|
1VDE
 
 | |
1TT9
 
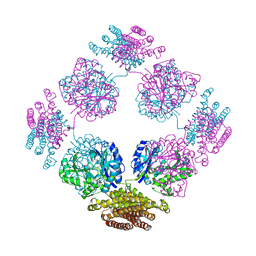 | | Structure of the bifunctional and Golgi associated formiminotransferase cyclodeaminase octamer | | Descriptor: | Formimidoyltransferase-cyclodeaminase (Formiminotransferase- cyclodeaminase) (FTCD) (58 kDa microtubule-binding protein) | | Authors: | Mao, Y, Vyas, N.K, Vyas, M.N, Chen, D.H, Ludtke, S.J, Chiu, W, Quiocho, F.A. | | Deposit date: | 2004-06-22 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structure of the bifunctional and Golgi-associated formiminotransferase cyclodeaminase octamer
Embo J., 23, 2004
|
|
1PBP
 
 | | FINE TUNING OF THE SPECIFICITY OF THE PERIPLASMIC PHOSPHATE TRANSPORT RECEPTOR: SITE-DIRECTED MUTAGENESIS, LIGAND BINDING, AND CRYSTALLOGRAPHIC STUDIES | | Descriptor: | PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Wang, Z, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1994-07-20 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fine tuning the specificity of the periplasmic phosphate transport receptor. Site-directed mutagenesis, ligand binding, and crystallographic studies.
J.Biol.Chem., 269, 1994
|
|
1EAM
 
 | | VACCINIA METHYLTRANSFERASE VP39 MUTANT (EC: 2.7.7.19) | | Descriptor: | PROTEIN (VP39), S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Hu, G, Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1999-01-26 | | Release date: | 1999-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1JW5
 
 | |
1ADS
 
 | |
1FQC
 
 | | CRYSTAL STRUCTURE OF MALTOTRIOTOL BOUND TO CLOSED-FORM MALTODEXTRIN BINDING PROTEIN | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-D-glucose | | Authors: | Duan, X, Hall, J.A, Nikaido, H, Quiocho, F.A. | | Deposit date: | 2000-09-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding.
J.Mol.Biol., 306, 2001
|
|
1FQB
 
 | | STRUCTURE OF MALTOTRIOTOL BOUND TO OPEN-FORM MALTODEXTRIN BINDING PROTEIN IN P2(1)CRYSTAL FORM | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-sorbitol | | Authors: | Duan, X, Hall, J.A, Nikaido, H, Quiocho, F.A. | | Deposit date: | 2000-09-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding.
J.Mol.Biol., 306, 2001
|
|
1FQA
 
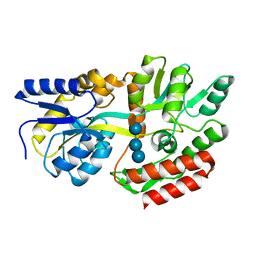 | | STRUCTURE OF MALTOTETRAITOL BOUND TO OPEN-FORM MALTODEXTRIN BINDING PROTEIN IN P2(1)CRYSTAL FORM | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-sorbitol | | Authors: | Duan, X, Hall, J.A, Nikaido, H, Quiocho, F.A. | | Deposit date: | 2000-09-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding.
J.Mol.Biol., 306, 2001
|
|
1JTF
 
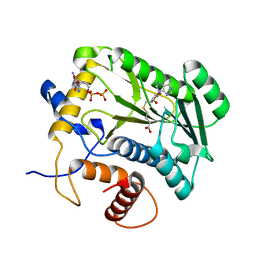 | | Crystal Structure Analysis of VP39-F180W mutant and m7GpppG complex | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Oguro, A, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 2001-08-20 | | Release date: | 2002-07-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The "cap-binding slot" of an mRNA cap-binding protein: quantitative effects of aromatic side chain choice in the double-stacking sandwich with cap.
Biochemistry, 41, 2002
|
|
1JSZ
 
 | | Crystal Structure Analysis of N7,9-dimethylguanine-VP39 complex | | Descriptor: | 7,9-DIMETHYLGUANINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Oguro, A, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 2001-08-19 | | Release date: | 2002-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The "cap-binding slot" of an mRNA cap-binding protein: quantitative effects of aromatic side chain choice in the double-stacking sandwich with cap.
Biochemistry, 41, 2002
|
|
1JTE
 
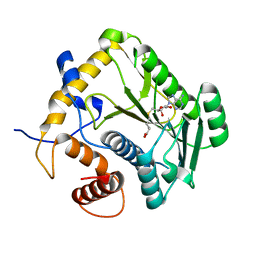 | | Crystal Structure Analysis of VP39 F180W mutant | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Oguro, A, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 2001-08-20 | | Release date: | 2002-07-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The "cap-binding slot" of an mRNA cap-binding protein: quantitative effects of aromatic side chain choice in the double-stacking sandwich with cap.
Biochemistry, 41, 2002
|
|
1JW4
 
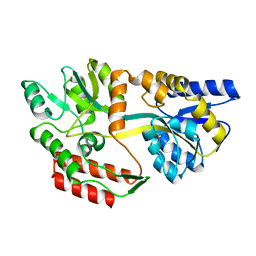 | | Structure of ligand-free maltodextrin-binding protein | | Descriptor: | maltodextrin-binding protein | | Authors: | Duan, X, Quiocho, F.A. | | Deposit date: | 2001-09-02 | | Release date: | 2002-01-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for a dominant role of nonpolar interactions in the binding of a transport/chemosensory receptor to its highly polar ligands.
Biochemistry, 41, 2002
|
|
1FQD
 
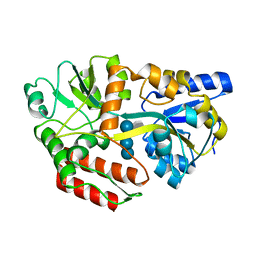 | | CRYSTAL STRUCTURE OF MALTOTETRAITOL BOUND TO CLOSED-FORM MALTODEXTRIN BINDING PROTEIN | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-D-glucose | | Authors: | Duan, X, Hall, J.A, Nikaido, H, Quiocho, F.A. | | Deposit date: | 2000-09-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding.
J.Mol.Biol., 306, 2001
|
|
1FRB
 
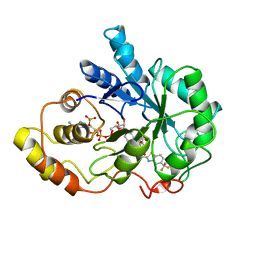 | | FR-1 PROTEIN/NADPH/ZOPOLRESTAT COMPLEX | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, FR-1 PROTEIN, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wilson, D.K, Quiocho, F.A. | | Deposit date: | 1995-08-08 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 A structure of FR-1, a fibroblast growth factor-induced member of the aldo-keto reductase family, complexed with coenzyme and inhibitor.
Biochemistry, 34, 1995
|
|
1B42
 
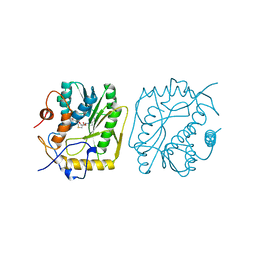 | | VACCINIA METHYLTRANSFERASE VP39 COMPLEXED WITH M1ADE AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 6-AMINO-1-METHYLPURINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1999-01-05 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1AV6
 
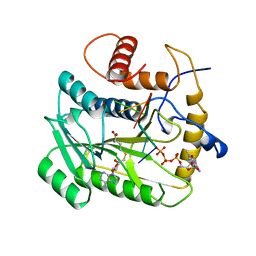 | | VACCINIA METHYLTRANSFERASE VP39 COMPLEXED WITH M7G CAPPED RNA HEXAMER AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, RNA (5'-R(*GP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1997-09-26 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for sequence-nonspecific recognition of 5'-capped mRNA by a cap-modifying enzyme.
Mol.Cell, 1, 1998
|
|
