2AS9
 
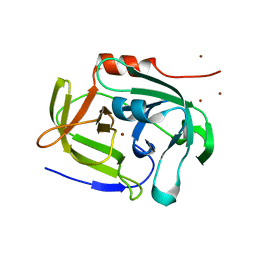 | | Functional and structural characterization of Spl proteases from staphylococcus aureus | | Descriptor: | ZINC ION, serine protease | | Authors: | Popowicz, G.M, Dubin, G, Stec-Niemczyk, J, Czarny, A, Dubin, A, Potempa, J, Holak, T.A. | | Deposit date: | 2005-08-23 | | Release date: | 2005-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional and Structural Characterization of Spl Proteases from Staphylococcus aureus
J.Mol.Biol., 358, 2006
|
|
1WLH
 
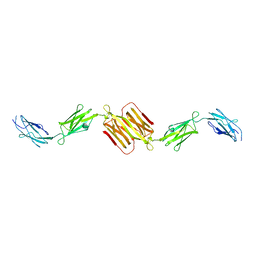 | | Molecular structure of the rod domain of Dictyostelium filamin | | Descriptor: | Gelation factor | | Authors: | Popowicz, G.M, Mueller, R, Noegel, A.A, Schleicher, M, Huber, R, Holak, T.A. | | Deposit date: | 2004-06-27 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular structure of the rod domain of dictyostelium filamin
J.Mol.Biol., 342, 2004
|
|
2Z5S
 
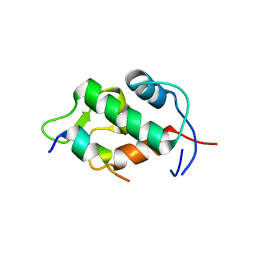 | | Molecular basis for the inhibition of p53 by Mdmx | | Descriptor: | Cellular tumor antigen p53, Mdm4 protein | | Authors: | Popowicz, G.M, Czarna, A, Rothweiler, U, Szwagierczak, A, Holak, T.A. | | Deposit date: | 2007-07-17 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the inhibition of p53 by Mdmx.
Cell Cycle, 6, 2007
|
|
2Z5T
 
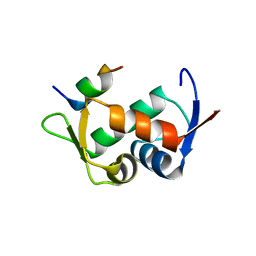 | | Molecular basis for the inhibition of p53 by Mdmx | | Descriptor: | Cellular tumor antigen p53, Mdm4 protein | | Authors: | Popowicz, G.M, Czarna, A, Rothweiler, U, Szwagierczak, A, Holak, T.A. | | Deposit date: | 2007-07-17 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the inhibition of p53 by Mdmx.
Cell Cycle, 6, 2007
|
|
3DAC
 
 | |
3DAB
 
 | |
3LBL
 
 | | Structure of human MDM2 protein in complex with Mi-63-analog | | Descriptor: | (2'R,3R,4'R,5'R)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-N-(2-morpholin-4-ylethyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
3LBK
 
 | | Structure of human MDM2 protein in complex with a small molecule inhibitor | | Descriptor: | 6-chloro-3-[1-(4-chlorobenzyl)-4-phenyl-1H-imidazol-5-yl]-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
3LBJ
 
 | | Structure of human MDMX protein in complex with a small molecule inhibitor | | Descriptor: | N-[(3S)-1-({6-chloro-3-[1-(4-chlorobenzyl)-4-phenyl-1H-imidazol-5-yl]-1H-indol-2-yl}carbonyl)pyrrolidin-3-yl]-N,N',N'-trimethylpropane-1,3-diamine, Protein Mdm4, SULFATE ION | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
8OS1
 
 | | X-ray structure of the Peroxisomal Targeting Signal 1 (PTS1) of Trypanosoma Cruzi PEX5 in complex with the PTS1 peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Napolitano, V, Blat, A, Popowicz, G.M, Dubin, G. | | Deposit date: | 2023-04-17 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural dynamics of the TPR domain of the peroxisomal cargo receptor Pex5 in Trypanosoma.
Int.J.Biol.Macromol., 280, 2024
|
|
4MVN
 
 | | Crystal structure of the staphylococcal serine protease SplA in complex with a specific phosphonate inhibitor | | Descriptor: | Serine protease splA, [(1S)-1-{[(benzyloxy)carbonyl]amino}-2-phenylethyl]phosphonic acid | | Authors: | Zdzalik, M, Burchacka, E, Niemczyk, J.S, Pustelny, K, Popowicz, G.M, Wladyka, B, Dubin, A, Potempa, J, Sienczyk, M, Dubin, G, Oleksyszyn, J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development and binding characteristics of phosphonate inhibitors of SplA protease from Staphylococcus aureus.
Protein Sci., 23, 2014
|
|
7R2V
 
 | | Structure of nsp14 from SARS-CoV-2 in complex with SAH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Proofreading exoribonuclease nsp14, ... | | Authors: | Czarna, A, Plewka, J, Kresik, L, Matsuda, A, Abdulkarim, K, Robinson, C, OByrne, S, Cunningham, F, Georgiou, I, Pachota, M, Popowicz, G.M, Wyatt, P.G, Dubin, G, Pyrc, K. | | Deposit date: | 2022-02-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Refolding of lid subdomain of SARS-CoV-2 nsp14 upon nsp10 interaction releases exonuclease activity.
Structure, 30, 2022
|
|
2ABZ
 
 | | Crystal structure of C19A/C43A mutant of leech carboxypeptidase inhibitor in complex with bovine carboxypeptidase A | | Descriptor: | Carboxypeptidase A1, Metallocarboxypeptidase inhibitor, ZINC ION | | Authors: | Arolas, J.L, Popowicz, G.M, Bronsoms, S, Aviles, F.X, Huber, R, Holak, T.A, Ventura, S. | | Deposit date: | 2005-07-18 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Study of a major intermediate in the oxidative folding of leech carboxypeptidase inhibitor: contribution of the fourth disulfide bond
J.Mol.Biol., 352, 2005
|
|
7QRC
 
 | | X-ray structure of Trypanosoma cruzi PEX14 in complex with a PEX5-PEX14 PPI inhibitor | | Descriptor: | GLYCEROL, Peroxin-14, ~{N}-(5-ethyl-6-oxidanylidene-benzo[b][1,4]benzothiazepin-2-yl)-2-(4-fluorophenyl)ethanamide | | Authors: | Napolitano, V, Popowicz, G.M, Dawidowski, M, Dubin, G. | | Deposit date: | 2022-01-10 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-based design, synthesis and evaluation of a novel family of PEX5-PEX14 interaction inhibitors against Trypanosoma.
Eur.J.Med.Chem., 243, 2022
|
|
5N8V
 
 | | Targeting the PEX14-PEX5 interaction by small molecules provides novel therapeutic routes to treat trypanosomiases. | | Descriptor: | 1-(2-azanylethyl)-5-[(4-methoxynaphthalen-1-yl)methyl]-~{N}-(naphthalen-1-ylmethyl)-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridine-3-carboxamide, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Dawidowski, M, Emmanouilidis, L, Sattler, M, Popowicz, G.M. | | Deposit date: | 2017-02-24 | | Release date: | 2017-03-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Inhibitors of PEX14 disrupt protein import into glycosomes and kill Trypanosoma parasites.
Science, 355, 2017
|
|
6YI4
 
 | | Structure of IMP-13 metallo-beta-lactamase complexed with citrate anion | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Zak, K.M, Zhou, R.X, Softley, C.A, Bostock, M.J, Sattler, M, Popowicz, G.M. | | Deposit date: | 2020-03-31 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of IMP-13 metallo-beta-lactamase complexed with citrate anion
Not published
|
|
5L87
 
 | | Targeting the PEX14-PEX5 interaction by small molecules provides novel therapeutic routes to treat trypanosomiases. | | Descriptor: | 1,2-ETHANEDIOL, 5-(1~{H}-indol-3-ylmethyl)-1-methyl-~{N}-(naphthalen-1-ylmethyl)-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridine-3-carboxamide, Peroxin 14 | | Authors: | Dawidowski, M, Emmanouilidis, L, Sattler, M, Popowicz, G.M. | | Deposit date: | 2016-06-07 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | Inhibitors of PEX14 disrupt protein import into glycosomes and kill Trypanosoma parasites.
Science, 355, 2017
|
|
5L8A
 
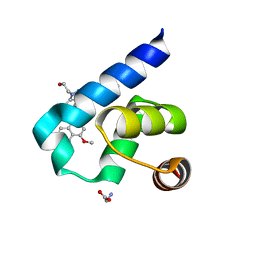 | | Targeting the PEX14-PEX5 interaction by small molecules provides novel therapeutic routes to treat trypanosomiases. | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-hydroxyethyl)-5-[(4-methoxynaphthalen-1-yl)methyl]-~{N}-(phenylmethyl)-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridine-3-carboxamide, GLYCINE, ... | | Authors: | Dawidowski, M, Emmanouilidis, L, Sattler, M, Popowicz, G.M. | | Deposit date: | 2016-06-07 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Inhibitors of PEX14 disrupt protein import into glycosomes and kill Trypanosoma parasites.
Science, 355, 2017
|
|
6I5P
 
 | |
6I68
 
 | |
6I7A
 
 | |
6I41
 
 | |
6F8G
 
 | |
6F8F
 
 | |
7NDX
 
 | | Crystal structure of the human HSP40 DNAJB1-CTDs in complex with a peptide of NudC | | Descriptor: | 1,2-ETHANEDIOL, DnaJ homolog subfamily B member 1, Nuclear migration protein nudC | | Authors: | Delhommel, F, Zak, K.M, Popowicz, G.M, Sattler, M. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | NudC guides client transfer between the Hsp40/70 and Hsp90 chaperone systems.
Mol.Cell, 82, 2022
|
|
