5IRR
 
 | | Crystal structure of Septin GTPase domain from Chlamydomonas reinhardtii | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Septin-like protein | | Authors: | Pinto, A.P.A, Pereira, H.M, Navarro, M.V.A.S, Brandao-Neto, J, Garratt, R.C, Araujo, A.P.U. | | Deposit date: | 2016-03-14 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Filaments and fingers: Novel structural aspects of the single septin from Chlamydomonas reinhardtii.
J. Biol. Chem., 292, 2017
|
|
3JBH
 
 | | TWO HEAVY MEROMYOSIN INTERACTING-HEADS MOTIFS FLEXIBLE DOCKED INTO TARANTULA THICK FILAMENT 3D-MAP ALLOWS IN DEPTH STUDY OF INTRA- AND INTERMOLECULAR INTERACTIONS | | Descriptor: | MYOSIN 2 ESSENTIAL LIGHT CHAIN STRIATED MUSCLE, MYOSIN 2 HEAVY CHAIN STRIATED MUSCLE, MYOSIN 2 REGULATORY LIGHT CHAIN STRIATED MUSCLE | | Authors: | Alamo, L, Qi, D, Wriggers, W, Pinto, A, Zhu, J, Bilbao, A, Gillilan, R.E, Hu, S, Padron, R. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Conserved Intramolecular Interactions Maintain Myosin Interacting-Heads Motifs Explaining Tarantula Muscle Super-Relaxed State Structural Basis.
J. Mol. Biol., 428, 2016
|
|
6OVD
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 3-ethylphenyl-ACEPC | | Descriptor: | (3S,5S)-5-[(2R)-2-amino-2-carboxyethyl]-1-(3-ethylphenyl)pyrazolidine-3-carboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Syrenne, J.T, Mou, T.C, Tamborini, L, Pinto, A, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 3-ethylphenyl-ACEPC
To Be Published
|
|
6OVE
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 4-propylphenyl-ACEPC | | Descriptor: | (3R,5S)-5-[(2R)-2-amino-2-carboxyethyl]-1-(4-propylphenyl)pyrazolidine-3-carboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Syrenne, J.T, Mou, T.C, Tamborini, L, Pinto, A, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 3-ethylphenyl-ACEPC
To Be Published
|
|
6GN0
 
 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, tetrameric crystal form | | Descriptor: | 14-3-3 protein beta/alpha, Exoenzyme S | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-29 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
6GNN
 
 | | Exoenzyme T from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, tetrameric crystal form bound to STO1101 | | Descriptor: | 14-3-3 protein beta/alpha, 3-(12-oxidanylidene-7-thia-9,11-diazatricyclo[6.4.0.0^{2,6}]dodeca-1(8),2(6),9-trien-10-yl)propanoic acid, Exoenzyme T | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
3JAX
 
 | | Heavy meromyosin from Schistosoma mansoni muscle thick filament by negative stain EM | | Descriptor: | myosin 2 heavy chain, myosin regulatory light chain, smooth muscle myosin essential light chain | | Authors: | Sulbaran, G, Alamo, L, Pinto, A, Marquez, G, Mendez, F, Padron, R, Craig, R. | | Deposit date: | 2015-07-03 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | An invertebrate smooth muscle with striated muscle myosin filaments.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6GNK
 
 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, trimeric crystal form bound to Carba-NAD | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 14-3-3 protein beta/alpha, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
6GNJ
 
 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, trimeric crystal form in complex with STO1101 | | Descriptor: | 14-3-3 protein beta/alpha, 3-(12-oxidanylidene-7-thia-9,11-diazatricyclo[6.4.0.0^{2,6}]dodeca-1(8),2(6),9-trien-10-yl)propanoic acid, Exoenzyme S | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
3DTP
 
 | | Tarantula heavy meromyosin obtained by flexible docking to Tarantula muscle thick filament Cryo-EM 3D-MAP | | Descriptor: | Myosin II regulatory light chain, Myosin light polypeptide 6, Myosin-11,Myosin-7 | | Authors: | Alamo, L, Wriggers, W, Pinto, A, Bartoli, F, Salazar, L, Zhao, F.Q, Craig, R, Padron, R. | | Deposit date: | 2008-07-15 | | Release date: | 2008-10-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Three-Dimensional Reconstruction of Tarantula Myosin Filaments Suggests How Phosphorylation May Regulate Myosin Activity
J.Mol.Biol., 384, 2008
|
|
4TVJ
 
 | | HUMAN ARTD2 (PARP2) - CATALYTIC DOMAIN IN COMPLEX WITH OLAPARIB | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Karlberg, T, Thorsell, A.G, Ekblad, T, Pinto, A.F, Schuler, H. | | Deposit date: | 2014-06-27 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J. Med. Chem., 60, 2017
|
|
4UND
 
 | | HUMAN ARTD1 (PARP1) - CATALYTIC DOMAIN IN COMPLEX WITH INHIBITOR TALAZOPARIB | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, POLY [ADP-RIBOSE] POLYMERASE 1, SODIUM ION | | Authors: | Karlberg, T, Thorsell, A.G, Ekblad, T, Klepsch, M, Pinto, A.F, Tresaugues, L, Moche, M, Schuler, H. | | Deposit date: | 2014-05-27 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J. Med. Chem., 60, 2017
|
|
5TBY
 
 | | HUMAN BETA CARDIAC HEAVY MEROMYOSIN INTERACTING-HEADS MOTIF OBTAINED BY HOMOLOGY MODELING (USING SWISS-MODEL) OF HUMAN SEQUENCE FROM APHONOPELMA HOMOLOGY MODEL (PDB-3JBH), RIGIDLY FITTED TO HUMAN BETA-CARDIAC NEGATIVELY STAINED THICK FILAMENT 3D-RECONSTRUCTION (EMD-2240) | | Descriptor: | Myosin light chain 3, Myosin regulatory light chain 2, ventricular/cardiac muscle isoform, ... | | Authors: | ALAMO, L, WARE, J.S, PINTO, A, GILLILAN, R.E, SEIDMAN, J.G, SEIDMAN, C.E, PADRON, R. | | Deposit date: | 2016-09-13 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes.
Elife, 6, 2017
|
|
5DEX
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, phenyl-ACEPC | | Descriptor: | 5-[(2R)-2-amino-2-carboxyethyl]-1-phenyl-1H-pyrazole-3-carboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Mou, T.-C, Conti, P, Pinto, A, Tamborini, L, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2015-08-26 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.
Proc. Natl. Acad. Sci. U.S.A., 2017
|
|
6HXR
 
 | | Human PARP16 (ARTD15) IN COMPLEX WITH EB-47 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Mono [ADP-ribose] polymerase PARP16 | | Authors: | Karlberg, T, Pinto, A.F, Thorsell, A.G, Schuler, H. | | Deposit date: | 2018-10-18 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Human PARP16 (ARTD15) IN COMPLEX WITH EB-47
To Be Published
|
|
6HXS
 
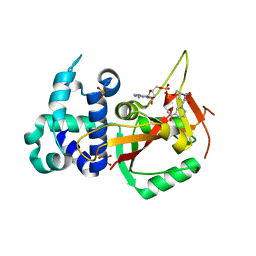 | | Human PARP16 (ARTD15) IN COMPLEX WITH CARBA-NAD | | Descriptor: | ADENOSINE, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Karlberg, T, Pinto, A.F, Thorsell, A.G, Schuler, H. | | Deposit date: | 2018-10-18 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Human PARP16 (ARTD15) IN COMPLEX WITH CARBA-NAD
To Be Published
|
|
6GN8
 
 | | Exoenzyme S from Pseudomonas aeruginosa in complex with human 14-3-3 protein beta, trimeric crystal form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 14-3-3 protein beta/alpha, Exoenzyme S | | Authors: | Karlberg, T, Pinto, A.F, Hornyak, P, Thorsell, A.G, Nareoja, K, Schuler, H. | | Deposit date: | 2018-05-30 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | 14-3-3 proteins activate Pseudomonas exotoxins-S and -T by chaperoning a hydrophobic surface.
Nat Commun, 9, 2018
|
|
4BK8
 
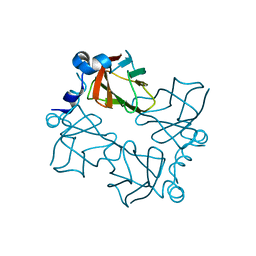 | | Superoxide reductase (Neelaredoxin) from Ignicoccus hospitalis | | Descriptor: | DESULFOFERRODOXIN, FERROUS IRON-BINDING REGION, FE (III) ION | | Authors: | Romao, C.V, Matias, P.M, Pinho, F.G, Sousa, C.M, Barradas, A.R, Pinto, A.F, Teixeira, M, Bandeiras, T.M. | | Deposit date: | 2013-04-22 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Structure of a Natural Sor Mutant
To be Published
|
|
5TSQ
 
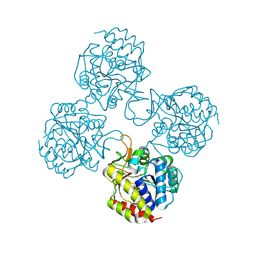 | | Crystal structure of IUnH from Leishmania braziliensis in complex with D-Ribose | | Descriptor: | CALCIUM ION, IUnH, beta-D-ribofuranose | | Authors: | Bachega, J.F.R, Timmers, L.F.S.M, Dalberto, P.F, Martinelli, L, Pinto, A.F.M, Basso, L.A, Norberto de Souza, O, Santos, D.S. | | Deposit date: | 2016-10-31 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structure of IUnH from Leishmania braziliensis in complex with D-Ribose
To Be Published
|
|
5VIH
 
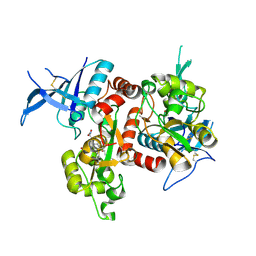 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 4-fluorophenyl-ACEPC | | Descriptor: | 5-[(2R)-2-amino-2-carboxyethyl]-1-(4-fluorophenyl)-1H-pyrazole-3-carboxylic acid, DI(HYDROXYETHYL)ETHER, GLYCINE, ... | | Authors: | Mou, T.-C, Conti, P, Pinto, A, Tamborini, L, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2017-04-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VIJ
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 4-bromophenyl-ACEPC | | Descriptor: | 5-[(2R)-2-amino-2-carboxyethyl]-1-(4-bromophenyl)-1H-pyrazole-3-carboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Mou, T.-C, Conti, P, Pinto, A, Tamborini, L, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2017-04-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VII
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 4-(3-fluoropropyl)phenyl-ACEPC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-[(2R)-2-amino-2-carboxyethyl]-1-[4-(3-fluoropropyl)phenyl]-1H-pyrazole-3-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mou, T.-C, Conti, P, Pinto, A, Tamborini, L, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2017-04-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4BFJ
 
 | | Superoxide reductase (Neelaredoxin) from Archaeoglobus fulgidus E12V mutant | | Descriptor: | FE (III) ION, SUPEROXIDE REDUCTASE | | Authors: | Bandeiras, T.M, Rodrigues, J.V, Sousa, C.M, Barradas, A.R, Pinho, F.G, Pinto, A.F, Teixeira, M, Matias, P.M, Romao, C.V. | | Deposit date: | 2013-03-19 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Understanding the Role of Key Residues in the Superoxide Reductase Molecular Mechanism, Exploring Archaeoglobus Fulgidus Sor Structure
To be Published
|
|
4C4U
 
 | | Superoxide reductase (Neelaredoxin) from Archaeoglobus fulgidus E12Q mutant in the reduced form | | Descriptor: | FE (II) ION, SUPEROXIDE REDUCTASE | | Authors: | Bandeiras, T.M, Rodrigues, J.V, Sousa, C.M, Barradas, A.R, Pinho, F.G, Pinto, A.F, Teixeira, M, Matias, P.M, Romao, C.V. | | Deposit date: | 2013-09-09 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.583 Å) | | Cite: | Understanding the Role of Key Residues in the Superoxide Reductase Molecular Mechanism, Exploring Archaeoglobus Fulgidus Sor Structure
To be Published
|
|
4C4B
 
 | | Superoxide reductase (Neelaredoxin) from Archaeoglobus fulgidus E12V in the reduced form | | Descriptor: | 1,2-ETHANEDIOL, FE (II) ION, SUPEROXIDE REDUCTASE | | Authors: | Bandeiras, T.M, Rodrigues, J.V, Sousa, C.M, Barradas, A.R, Pinho, F.G, Pinto, A.F, Teixeira, M, Matias, P.M, Romao, C.V. | | Deposit date: | 2013-09-03 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Understanding the Role of Key Residues in the Superoxide Reductase Molecular Mechanism, Exploring Archaeoglobus Fulgidus Sor Structure
To be Published
|
|
