3GOE
 
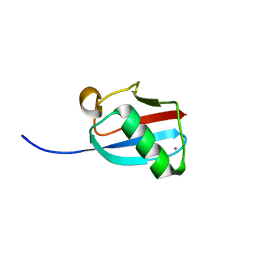 | | Molecular Mimicry of SUMO promotes DNA repair | | Descriptor: | CALCIUM ION, DNA repair protein rad60 | | Authors: | Perry, J.J.P. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Molecular mimicry of SUMO promotes DNA repair.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2P4K
 
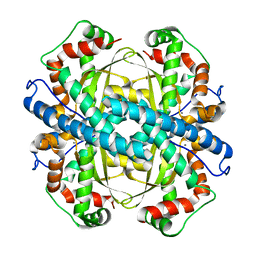 | |
4PPE
 
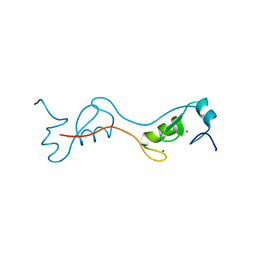 | | human RNF4 RING domain | | Descriptor: | E3 ubiquitin-protein ligase RNF4, ZINC ION | | Authors: | Perry, J.J, Arvai, A.S, Hitomi, C, Tainer, J.A. | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RNF4 interacts with both SUMO and nucleosomes to promote the DNA damage response.
Embo Rep., 15, 2014
|
|
6UMQ
 
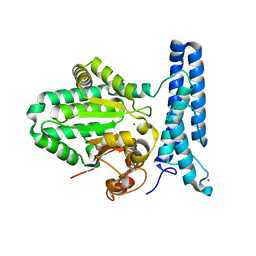 | | Structure of DUF89 | | Descriptor: | Damage-control phosphatase DUF89, MAGNESIUM ION | | Authors: | Perry, J.J, Kenjic, N, Dennis, T.N. | | Deposit date: | 2019-10-10 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Human ARMT1 structure and substrate specificity indicates that it is a DUF89 family damage-control phosphatase.
J.Struct.Biol., 212, 2020
|
|
6UMR
 
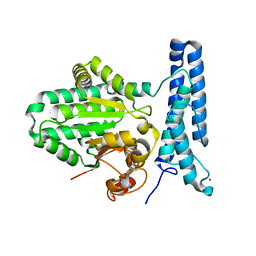 | | Structure of DUF89 - D291A mutant | | Descriptor: | Damage-control phosphatase DUF89, MAGNESIUM ION | | Authors: | Perry, J.J, Kenjic, N, Dennis, T.N. | | Deposit date: | 2019-10-10 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Human ARMT1 structure and substrate specificity indicates that it is a DUF89 family damage-control phosphatase.
J.Struct.Biol., 212, 2020
|
|
2FBX
 
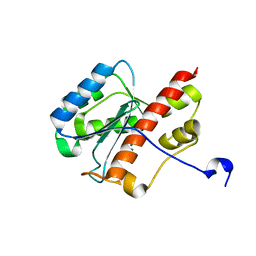 | | WRN exonuclease, Mg complex | | Descriptor: | MAGNESIUM ION, Werner syndrome helicase | | Authors: | Perry, J.J. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2FC0
 
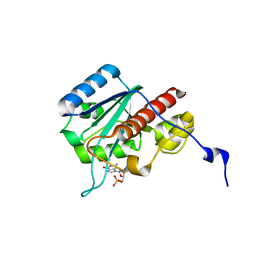 | | WRN exonuclease, Mn dGMP complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, Werner syndrome helicase | | Authors: | Perry, J.J, Tainer, J.A. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2FBY
 
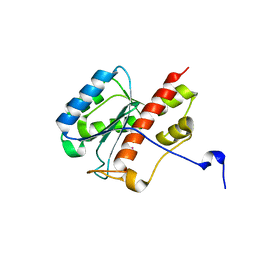 | | WRN exonuclease, Eu complex | | Descriptor: | EUROPIUM (III) ION, Werner syndrome helicase | | Authors: | Perry, J.J. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2FBV
 
 | | WRN exonuclease, Mn complex | | Descriptor: | MANGANESE (II) ION, Werner syndrome helicase | | Authors: | Perry, J.J. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2FBT
 
 | | WRN exonuclease | | Descriptor: | ACETIC ACID, Werner syndrome helicase | | Authors: | Perry, J.J. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
3HVC
 
 | | Crystal structure of human p38alpha MAP kinase | | Descriptor: | 4-[3-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL]PYRIDINE, Mitogen-activated protein kinase 14 | | Authors: | Perry, J.J, Tainer, J.A. | | Deposit date: | 2009-06-15 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | p38alpha MAP kinase C-terminal domain binding pocket characterized by crystallographic and computational analyses.
J.Mol.Biol., 391, 2009
|
|
3RCZ
 
 | | Rad60 SLD2 Ubc9 Complex | | Descriptor: | DNA repair protein rad60, SUMO-conjugating enzyme ubc9 | | Authors: | Perry, J.J.P, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2011-03-31 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA repair and global sumoylation are regulated by distinct Ubc9 noncovalent complexes.
Mol.Cell.Biol., 31, 2011
|
|
3RD2
 
 | | NIP45 SUMO-like Domain 2 | | Descriptor: | NFATC2-interacting protein | | Authors: | Perry, J.J.P, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2011-03-31 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA repair and global sumoylation are regulated by distinct Ubc9 noncovalent complexes.
Mol.Cell.Biol., 31, 2011
|
|
2GDS
 
 | |
6VBX
 
 | | Crystal structure of Mcl-1 in complex with 138E12 peptide, Lys-covalent antagonist | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, Synthetic peptide | | Authors: | Pellecchia, M, Perry, J.J, Kenjic, N, Assar, Z. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, Synthesis, and Structural Characterization of Lysine Covalent BH3 Peptides Targeting Mcl-1.
J.Med.Chem., 64, 2021
|
|
6WML
 
 | | Human TLR8 bound to the potent agonist, GS-9688 (Selgantolimod) | | Descriptor: | (2R)-2-[(2-amino-7-fluoropyrido[3,2-d]pyrimidin-4-yl)amino]-2-methylhexan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 8, ... | | Authors: | Appleby, T.C, Perry, J.K, Mish, M, Villasenor, A.G, Mackman, R.L. | | Deposit date: | 2020-04-21 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of GS-9688 (Selgantolimod) as a Potent and Selective Oral Toll-Like Receptor 8 Agonist for the Treatment of Chronic Hepatitis B.
J.Med.Chem., 63, 2020
|
|
3MML
 
 | | Allophanate Hydrolase Complex from Mycobacterium smegmatis, Msmeg0435-Msmeg0436 | | Descriptor: | Allophanate hydrolase subunit 1, Allophanate hydrolase subunit 2, CHLORIDE ION | | Authors: | Kaufmann, M, Chernishof, I, Shin, A, Germano, D, Sawaya, M.R, Waldo, G.S, Arbing, M.A, Perry, J, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Allphanate Hydrolase Complex from M. smegmatis, Msmeg0435-Msmeg0436
To be Published
|
|
4O24
 
 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | (5~{Z})-5-[(4-hydroxyphenyl)methylidene]-3-(2-methylpropyl)-2-sulfanylidene-1,3-thiazolidin-4-one, Exonuclease, putative, ... | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-16 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
4NZV
 
 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | Exonuclease, putative, MANGANESE (II) ION | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-12 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
8GL9
 
 | | Co-crystal structure of caPCNA bound to AOH1160 derivative 1LE | | Descriptor: | CHLORIDE ION, N~2~-(naphthalene-1-carbonyl)-N-(2-phenoxyphenyl)-L-alpha-glutamine, Proliferating cell nuclear antigen | | Authors: | Jossart, J, Kenjic, N, Malkas, L.H, Hickey, R.J, Perry, J.J. | | Deposit date: | 2023-03-21 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Small molecule targeting of transcription-replication conflict for selective chemotherapy.
Cell Chem Biol, 30, 2023
|
|
8GLA
 
 | |
4O4K
 
 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | (5~{E})-2-azanylidene-5-[(4-hydroxyphenyl)methylidene]-1,3-thiazolidin-4-one, Exonuclease, putative, ... | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
4O43
 
 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | (5~{E})-3-[(2~{R})-butan-2-yl]-5-[(4-hydroxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, Exonuclease, putative, ... | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
4O5G
 
 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | (5~{E})-5-[(4-aminophenyl)methylidene]-2-azanylidene-1,3-thiazolidin-4-one, Exonuclease, putative, ... | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
6RJP
 
 | | Bfl-1 in complex with alpha helical peptide | | Descriptor: | Bcl-2-like protein 11, Bcl-2-related protein A1 | | Authors: | Baggio, C, Gambini, L, Udompholkul, P, Salem, A.F, Hakansson, M, Jossart, J, Perry, J, Pellecchia, M. | | Deposit date: | 2019-04-29 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | N-locking stabilization of covalent helical peptides: Application to Bfl-1 antagonists.
Chem.Biol.Drug Des., 95, 2020
|
|
