7B80
 
 | | DeAMPylation complex of monomeric FICD and AMPylated BiP (state 2) | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Perera, L.A, Ron, D. | | Deposit date: | 2020-12-12 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structures of a deAMPylation complex rationalise the switch between antagonistic catalytic activities of FICD.
Nat Commun, 12, 2021
|
|
7B7Z
 
 | | DeAMPylation complex of monomeric FICD and AMPylated BiP (state 1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Endoplasmic reticulum chaperone BiP, ... | | Authors: | Perera, L.A, Ron, D. | | Deposit date: | 2020-12-12 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of a deAMPylation complex rationalise the switch between antagonistic catalytic activities of FICD.
Nat Commun, 12, 2021
|
|
7A4V
 
 | |
7A4U
 
 | |
6I7G
 
 | | Crystal structure of dimeric wild type FICD complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
6I7L
 
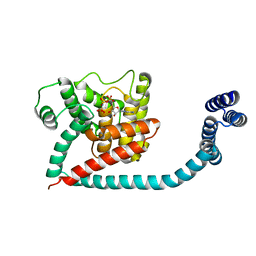 | | Crystal structure of monomeric FICD mutant L258D complexed with MgAMP-PNP | | Descriptor: | Adenosine monophosphate-protein transferase FICD, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
6I7K
 
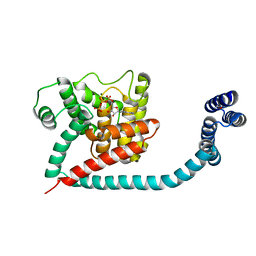 | | Crystal structure of monomeric FICD mutant L258D complexed with MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, ETHANOL, ... | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
6I7J
 
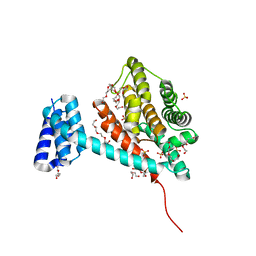 | | Crystal structure of monomeric FICD mutant L258D | | Descriptor: | Adenosine monophosphate-protein transferase FICD, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
6I7I
 
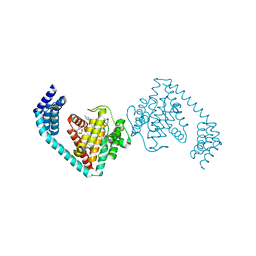 | | Crystal structure of dimeric FICD mutant K256A complexed with MgATP | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ADENOSINE-5'-TRIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, ... | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
6I7H
 
 | | Crystal structure of dimeric FICD mutant K256S | | Descriptor: | Adenosine monophosphate-protein transferase FICD, MAGNESIUM ION | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
6MW7
 
 | | Crystal structure of ATPase module of SMCHD1 bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Pedersen, L.C, Inoue, K, Kim, S, Perera, L, Shaw, N.D. | | Deposit date: | 2018-10-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | A ubiquitin-like domain is required for stabilizing the N-terminal ATPase module of human SMCHD1.
Commun Biol, 2, 2019
|
|
4ZCE
 
 | | Crystal Structure of the dust mite allergen Der p 23 from Dermatophagoides pteronyssinus | | Descriptor: | 1,2-ETHANEDIOL, Dust mite allergen | | Authors: | Pedersen, L.C, Mueller, G.A, Randall, T.A, Glesner, J, Perera, L, Edwards, L.L, Chapman, M.D, London, R.E, Pomes, A. | | Deposit date: | 2015-04-15 | | Release date: | 2015-11-25 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Serological, genomic and structural analyses of the major mite allergen Der p 23.
Clin Exp Allergy, 46, 2016
|
|
4OUO
 
 | | anti-Bla g 1 scFv | | Descriptor: | CHLORIDE ION, SULFATE ION, anti Bla g 1 scFv | | Authors: | Mueller, G.A, Ankney, J.A, Glesner, J, Khurana, T, Edwards, L.L, Pedersen, L.C, Perera, L, Slater, J.E, Pomes, A, London, R.E. | | Deposit date: | 2014-02-18 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of an anti-Bla g 1 scFv: Epitope mapping and cross-reactivity.
Mol.Immunol., 59, 2014
|
|
4JWN
 
 | | Ternary complex of D256A mutant of DNA Polymerase Beta | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Perera, L, Ping, L, Shock, D.D, Beard, W.A, Pedersen, L.C, Pedersen, L.G, Wilson, S.H. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Amino Acid Substitution in the Active Site of DNA Polymerase beta Explains the Energy Barrier of the Nucleotidyl Transfer Reaction.
J.Am.Chem.Soc., 135, 2013
|
|
4JWM
 
 | | Ternary complex of D256E mutant of DNA Polymerase Beta | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Perera, L, Ping, L, Shock, D.D, Beard, W.A, Pedersen, L.C, Pedersen, L.G, Wilson, S.H. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Amino Acid Substitution in the Active Site of DNA Polymerase beta Explains the Energy Barrier of the Nucleotidyl Transfer Reaction.
J.Am.Chem.Soc., 135, 2013
|
|
4NDZ
 
 | | Structure of Maltose Binding Protein fusion to 2-O-Sulfotransferase with bound heptasaccharide and PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Maltose-binding periplasmic protein, Heparan sulfate 2-O-sulfotransferase 1 fusion, ... | | Authors: | Liu, C, Sheng, J, Krahn, J.M, Perera, L, Xu, Y, Hsieh, P, Liu, J, Pedersen, L.C. | | Deposit date: | 2013-10-28 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Deciphering the role of 2-O-sulfotransferase in regulating heparan sulfate biosynthesis
To be Published
|
|
3HX0
 
 | | ternary complex of L277A, H511A, R514 mutant pol lambda bound to a 2 nucleotide gapped DNA substrate with a scrunched dA | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*AP*GP*TP*AP*T)-3', 5'-D(*CP*GP*GP*CP*AP*AP*AP*TP*AP*CP*TP*G)-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Larrea, A.A, Havener, J.M, Perera, L, Krahn, J.M, Pedersen, L.C, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2009-06-19 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Scrunching During DNA Repair Synthesis
To be Published
|
|
3RH5
 
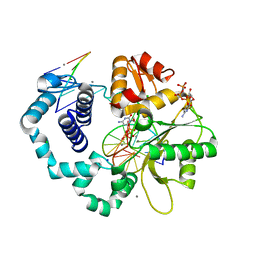 | | DNA Polymerase Beta Mutant (Y271) with a dideoxy-terminated primer with an incoming deoxynucleotide (dCTP) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*GP*CP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DDG))-3', ... | | Authors: | Cavanaugh, N.A, Beard, W.A, Batra, V.K, Perera, L, Pedersen, L.G, Wilson, S.H. | | Deposit date: | 2011-04-11 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Molecular insights into DNA polymerase deterrents for ribonucleotide insertion.
J.Biol.Chem., 286, 2011
|
|
3RH4
 
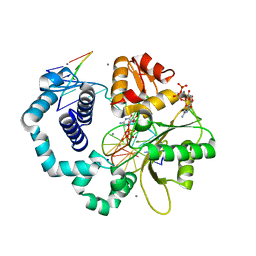 | | DNA Polymerase Beta with a dideoxy-terminated primer with an incoming ribonucleotide (rCTP) | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*GP*CP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DDG))-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Cavanaugh, N.A, Beard, W.A, Batra, V.K, Perera, L, Pedersen, L.G, Wilson, S.H. | | Deposit date: | 2011-04-11 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.918 Å) | | Cite: | Molecular insights into DNA polymerase deterrents for ribonucleotide insertion.
J.Biol.Chem., 286, 2011
|
|
3RH6
 
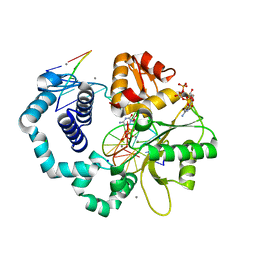 | | DNA Polymerase Beta Mutant (Y271) with a dideoxy-terminated primer with an incoming ribonucleotide (rCTP) | | Descriptor: | (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DDG))-3'), (5'-D(P*GP*TP*CP*GP*G)-3'), 5'-D(*CP*CP*GP*AP*CP*GP*CP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', ... | | Authors: | Cavanaugh, N.A, Beard, W.A, Batra, V.K, Perera, L, Pedersen, L.G, Wilson, S.H. | | Deposit date: | 2011-04-11 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | Molecular insights into DNA polymerase deterrents for ribonucleotide insertion.
J.Biol.Chem., 286, 2011
|
|
3HW8
 
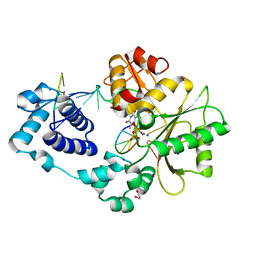 | | ternary complex of DNA polymerase lambda of a two nucleotide gapped DNA substrate with a C in the scrunch site | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*AP*GP*TP*AP*T)-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Larrea, A.A, Havener, J.M, Perera, L, Krahn, J.M, Pedersen, L.C, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2009-06-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Scrunching Durning DNA Repair Synthesis
To be Published
|
|
2L7D
 
 | | Ribonucleotide Perturbation of DNA Structure: Solution Structure of [d(CGC)r(G)d(AATTCGCG)]2 | | Descriptor: | 5'-D(*CP*GP*C)-R(P*G)-D(P*AP*AP*TP*TP*CP*GP*CP*G)-3' | | Authors: | DeRose, E.F, Perera, L, Murray, M.S, Kunkel, T.A, London, R.E. | | Deposit date: | 2010-12-07 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Dickerson DNA dodecamer containing a single ribonucleotide.
Biochemistry, 51, 2012
|
|
