6TYT
 
 | |
7M0E
 
 | | Pre-catalytic synaptic complex of DNA Polymerase Lambda with gapped DSB substrate and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*GP*C)-3'), ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
7M09
 
 | | Pre-catalytic quaternary complex of DNA Polymerase Lambda with blunt-ended DSB substrate and incoming dUMPNPP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
7M0D
 
 | | Pre-catalytic quaternary complex of DNA Polymerase Lambda with bound complementary DSB substrate and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
7M0A
 
 | | Incomplete in crystallo incorporation by DNA Polymerase Lambda bound to blunt-ended DSB substrate and incoming dTTP | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*CP*AP*GP*TP*GP*CP*T)-3'), ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
7M0B
 
 | | Pre-catalytic quaternary complex of DNA Polymerase Lambda with bound mismatched DSB and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
7M08
 
 | | Post-catalytic nicked complex of DNA Polymerase Lambda with bound 1-nt gapped SSB substrate and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comprehensive structural survey of DNA double-strand break synapsis by DNA Polymerase Lambda
Not Published
|
|
7M0C
 
 | | Post-catalytic nicked complex of DNA Polymerase Lambda with bound mismatched DSB substrate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*AP*GP*TP*GP*CP*T)-3'), ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Comprehensive structural survey of DNA double-strand break synapsis by DNA Polymerase Lambda
Not Published
|
|
7M07
 
 | | Pre-catalytic ternary complex of DNA Polymerase Lambda with bound 1-nt gapped SSB substrate and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
1AQY
 
 | | ESTROGEN SULFOTRANSFERASE WITH PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, ESTROGEN SULFOTRANSFERASE | | Authors: | Kakuta, Y, Pedersen, L.G, Carter, C.W, Negishi, M, Pedersen, L.C. | | Deposit date: | 1997-08-04 | | Release date: | 1998-10-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of estrogen sulphotransferase.
Nat.Struct.Biol., 4, 1997
|
|
8EPU
 
 | |
8EPV
 
 | | 2.2 A crystal structure of the lipocalin cat allergen Fel d 7 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Fel d 7 allergen, ... | | Authors: | Min, J, Pedersen, L.C, Geoffrey, M.A. | | Deposit date: | 2022-10-06 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural and ligand binding analysis of the pet allergens Can f 1 and Fel d 7.
Front Allergy, 4, 2023
|
|
7SCD
 
 | | Ternary complex of fixed-arm Trx-3ost5 (I299E) with 8mer-1 octasaccharide substrate and co-factor product PAP | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, Thioredoxin 1,Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Wander, R, Kaminski, A.M, Krahn, J.M, Liu, J, Pedersen, L.C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Substrate Specificity Analysis of 3-O-Sulfotransferase Isoform 5 to Synthesize Heparan Sulfate
Acs Catalysis, 11, 2021
|
|
7SCE
 
 | | Ternary complex of fixed-arm Trx-3ost5 (I299E) with 8mer-2 octasaccharide substrate and co-factor product PAP | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, Thioredoxin 1,Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Wander, R, Kaminski, A.M, Krahn, J.M, Liu, J, Pedersen, L.C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and Substrate Specificity Analysis of 3-O-Sulfotransferase Isoform 5 to Synthesize Heparan Sulfate
Acs Catalysis, 11, 2021
|
|
5FGW
 
 | | Structure of Sda1 nuclease with bound zinc ion | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Moon, A.F, Krahn, J.M, Xun, L, Cuneo, M.J, Pedersen, L.C. | | Deposit date: | 2015-12-21 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of the virulence factor Sda1 nuclease from Streptococcus pyogenes.
Nucleic Acids Res., 44, 2016
|
|
5FGU
 
 | | Structure of Sda1 nuclease apoprotein as an EGFP fixed-arm fusion | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Green fluorescent protein,Extracellular streptodornase D, ... | | Authors: | Moon, A.F, Krahn, J.M, Xun, L, Cuneo, M.J, Pedersen, L.C. | | Deposit date: | 2015-12-21 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Structural characterization of the virulence factor Sda1 nuclease from Streptococcus pyogenes.
Nucleic Acids Res., 44, 2016
|
|
4JWN
 
 | | Ternary complex of D256A mutant of DNA Polymerase Beta | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Perera, L, Ping, L, Shock, D.D, Beard, W.A, Pedersen, L.C, Pedersen, L.G, Wilson, S.H. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Amino Acid Substitution in the Active Site of DNA Polymerase beta Explains the Energy Barrier of the Nucleotidyl Transfer Reaction.
J.Am.Chem.Soc., 135, 2013
|
|
4JWM
 
 | | Ternary complex of D256E mutant of DNA Polymerase Beta | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Perera, L, Ping, L, Shock, D.D, Beard, W.A, Pedersen, L.C, Pedersen, L.G, Wilson, S.H. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Amino Acid Substitution in the Active Site of DNA Polymerase beta Explains the Energy Barrier of the Nucleotidyl Transfer Reaction.
J.Am.Chem.Soc., 135, 2013
|
|
3LK9
 
 | | DNA polymerase beta with a gapped DNA substrate and dTMP(CF2)P(CF2)P | | Descriptor: | 5'-O-[(R)-{[(R)-[difluoro(phosphono)methyl](hydroxy)phosphoryl](difluoro)methyl}(hydroxy)phosphoryl]thymidine, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Zibinsky, M, Surya Prakash, G.K, Upton, T.G, Kashemirov, B.A, McKenna, C.E, Oertell, K, Goodman, M.F, Batra, V.K, Pedersen, L.C, Beard, W.A, Wilson, S.H. | | Deposit date: | 2010-01-27 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synthesis and biological evaluation of fluorinated deoxynucleotide analogs based on bis-(difluoromethylene)triphosphoric acid.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MBY
 
 | | Ternary complex of DNA Polymerase BETA with template base A and 8oxodGTP in the active site with a dideoxy terminated primer | | Descriptor: | 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Hou, E.W, Pedersen, L.C, Prasad, R, Wilson, S.H. | | Deposit date: | 2010-03-26 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenic conformation of 8-oxo-7,8-dihydro-2'-dGTP in the confines of a DNA polymerase active site.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MQ1
 
 | | Crystal Structure of Dust Mite Allergen Der p 5 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Mite allergen Der p 5, ... | | Authors: | Mueller, G.A, Gosavi, R.A, Krahn, J.M, Edwards, L.L, Cuneo, M.J, Glesner, J, Pomes, A, Chapman, M.D, London, R.E, Pedersen, L.C. | | Deposit date: | 2010-04-27 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Der p 5 crystal structure provides insight into the group 5 dust mite allergens.
J.Biol.Chem., 285, 2010
|
|
3OB4
 
 | | MBP-fusion protein of the major peanut allergen Ara h 2 | | Descriptor: | CHLORIDE ION, Maltose ABC transporter periplasmic protein,Arah 2, SULFATE ION, ... | | Authors: | Mueller, G.A, Gosavi, R.A, Moon, A.F, London, R.E, Pedersen, L.C. | | Deposit date: | 2010-08-06 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | Ara h 2: crystal structure and IgE binding distinguish two subpopulations of peanut allergic patients by epitope diversity.
Allergy, 66, 2011
|
|
6WIE
 
 | | Post-catalytic nicked complex of human Polymerase Mu on a complementary DNA double-strand break substrate | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*AP*CP*G)-3'), ... | | Authors: | Kaminski, A.M, Kunkel, T.A, Pedersen, L.C, Bebenek, K. | | Deposit date: | 2020-04-09 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural snapshots of human DNA polymerase mu engaged on a DNA double-strand break.
Nat Commun, 11, 2020
|
|
6WIC
 
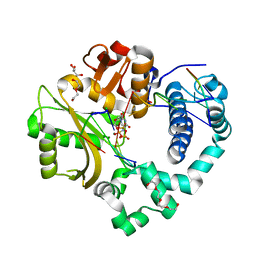 | | Pre-catalytic quaternary complex of human Polymerase Mu on a complementary DNA double-strand break substrate | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kaminski, A.M, Kunkel, T.A, Pedersen, L.C, Bebenek, K. | | Deposit date: | 2020-04-09 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural snapshots of human DNA polymerase mu engaged on a DNA double-strand break.
Nat Commun, 11, 2020
|
|
6WID
 
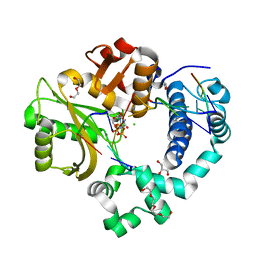 | | Nucleotide incorporation intermediate into quaternary complex of human Polymerase Mu on a complementary DNA double-strand break substrate | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kaminski, A.M, Kunkel, T.A, Pedersen, L.C, Bebenek, K. | | Deposit date: | 2020-04-09 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural snapshots of human DNA polymerase mu engaged on a DNA double-strand break.
Nat Commun, 11, 2020
|
|
