7SHP
 
 | | Crystal structure of hSTING in complex with c[2',3'-(ribo-2'-G, xylo-3'-A)-MP](RJ244) | | Descriptor: | (2S,5R,7R,8R,10S,12aR,14R,15R,15aR,16R)-7-(2-amino-6-oxo-3,6-dihydro-9H-purin-9-yl)-14-(6-amino-9H-purin-9-yl)-2,10,15,16-tetrahydroxyoctahydro-2H,10H,12H-5,8-methano-2lambda~5~,10lambda~5~-furo[3,2-l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Xie, W, Lama, L, Yang, X.J, Kuryavyi, V, Nudelman, I, Glickman, J.F, Jones, R.A, Tuschl, T, Patel, D.J. | | Deposit date: | 2021-10-11 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Arabinose- and xylose-modified analogs of 2',3'-cGAMP act as STING agonists.
Cell Chem Biol, 2023
|
|
1UOW
 
 | | Calcium binding domain C2B | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Cheng, Y, Sequeira, S.M, Sollner, T.H, Patel, D.J. | | Deposit date: | 2003-09-24 | | Release date: | 2004-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystallographic Identification of Ca2+ and Sr2+ Coordination Sites in Synaptotagmin I C2B Domain
Protein Sci., 13, 2004
|
|
1AM0
 
 | | AMP RNA APTAMER COMPLEX, NMR, 8 STRUCTURES | | Descriptor: | ADENOSINE MONOPHOSPHATE, RNA APTAMER | | Authors: | Jiang, F, Kumar, R.A, Jones, R.A, Patel, D.J. | | Deposit date: | 1997-06-19 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of RNA Folding and Recognition in an AMP-RNA Aptamer Complex
Nature, 382, 1996
|
|
4K85
 
 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 12:0 Ceramide-1-Phosphate (12:0-C1P) | | Descriptor: | (2S,3R,4E)-2-(dodecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-17 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
4K8N
 
 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 18:1 Ceramide-1-Phosphate (18:1-C1P) | | Descriptor: | (2S,3R,4Z)-3-hydroxy-2-[(9E)-octadec-9-enoylamino]octadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-18 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
1UOV
 
 | | Calcium binding domain C2B | | Descriptor: | CALCIUM ION, GLYCEROL, SYNAPTOTAGMIN I | | Authors: | Cheng, Y, Sequeira, S.M, Sollner, T.H, Patel, D.J. | | Deposit date: | 2003-09-24 | | Release date: | 2004-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallographic Identification of Ca2+ and Sr2+ Coordination Sites in Synaptotagmin I C2B Domain
Protein Sci., 13, 2004
|
|
1FJB
 
 | | NMR Study of an 11-Mer DNA Duplex Containing 7,8-Dihydro-8-Oxoadenine (AOXO) Opposite Thymine | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*(A38)P*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*TP*GP*TP*AP*CP*G)-3') | | Authors: | Chen, H, Johnson, F, Grollman, A.P, Patel, D.J. | | Deposit date: | 1995-12-15 | | Release date: | 1996-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Studies of the Ionizing Radiation Adduct 7,8-Dihydro-8-Oxoadenine (A Oxo) Positioned Opposite Thymine and Guanine in DNA Duplexes
MAGN.RESON.CHEM., 34, 1996
|
|
7R76
 
 | | Cryo-EM structure of DNMT5 in apo state | | Descriptor: | DNA repair protein Rad8, ZINC ION | | Authors: | Wang, J, Patel, D.J. | | Deposit date: | 2021-06-24 | | Release date: | 2022-02-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into DNMT5-mediated ATP-dependent high-fidelity epigenome maintenance.
Mol.Cell, 82, 2022
|
|
1FMN
 
 | | SOLUTION STRUCTURE OF FMN-RNA APTAMER COMPLEX, NMR, 5 STRUCTURES | | Descriptor: | FLAVIN MONONUCLEOTIDE, RNA (5'-R(*GP*GP*CP*GP*UP*GP*UP*AP*GP*GP *AP*UP*AP*UP*GP*CP*UP*UP*CP*GP*GP*CP*AP*GP*AP*AP*GP *GP*AP*CP*AP*CP*GP*CP*C)-3') | | Authors: | Fan, P, Suri, A.K, Fiala, R, Live, D, Patel, D.J. | | Deposit date: | 1995-12-04 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular recognition in the FMN-RNA aptamer complex.
J.Mol.Biol., 258, 1996
|
|
1Y27
 
 | | G-riboswitch-guanine complex | | Descriptor: | Bacillus subtilis xpt, GUANINE | | Authors: | Serganov, A, Yuan, Y.R, Patel, D.J. | | Deposit date: | 2004-11-20 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Discriminative Regulation of Gene Expression by Adenine- and Guanine-Sensing mRNAs
Chem.Biol., 11, 2004
|
|
1AW4
 
 | |
1BIV
 
 | | BOVINE IMMUNODEFICIENCY VIRUS TAT-TAR COMPLEX, NMR, 5 STRUCTURES | | Descriptor: | TAR RNA, TAT PEPTIDE | | Authors: | Ye, X, Kumar, R.A, Patel, D.J. | | Deposit date: | 1996-06-12 | | Release date: | 1996-12-23 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Molecular recognition in the bovine immunodeficiency virus Tat peptide-TAR RNA complex.
Chem.Biol., 2, 1995
|
|
4KF6
 
 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 8:0 Ceramide-1-Phosphate (8:0-C1P) | | Descriptor: | (2S,3R,4E)-3-hydroxy-2-(octanoylamino)octadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-26 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.195 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
4QUF
 
 | | crystal structure of chromodomain of Rhino with H3K9me3 | | Descriptor: | H3(1-15)K9me3 peptide, RE36324p | | Authors: | Li, S, Patel, D.J. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing.
Genes Dev., 28, 2014
|
|
4RGF
 
 | | Crystal structure of the in-line aligned env22 twister ribozyme soaked with Mn2+ | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Ren, A, Rajashankar, K.R, Simanshu, D, Patel, D. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2008 Å) | | Cite: | In-line alignment and Mg(2+) coordination at the cleavage site of the env22 twister ribozyme.
Nat Commun, 5, 2014
|
|
4RGE
 
 | | Crystal structure of the in-line aligned env22 twister ribozyme | | Descriptor: | MAGNESIUM ION, env22 twister ribozyme | | Authors: | Ren, A, Rajashankar, K.R, Simanshu, D, Patel, D. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | In-line alignment and Mg(2+) coordination at the cleavage site of the env22 twister ribozyme.
Nat Commun, 5, 2014
|
|
4K9B
 
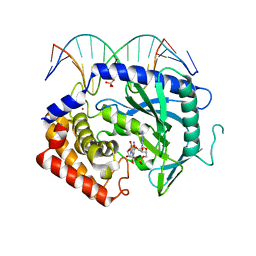 | | Structure of Ternary Complex of cGAS with dsDNA and Bound c[G(2 ,5 )pA(3 ,5 )p] | | Descriptor: | ADENOSINE MONOPHOSPHATE, Cyclic GMP-AMP synthase, DNA-F, ... | | Authors: | Gao, P, Wu, Y, Patel, D.J. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase.
Cell(Cambridge,Mass.), 153, 2013
|
|
1F3S
 
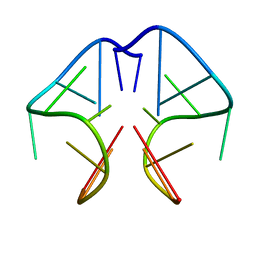 | | Solution Structure of DNA Sequence GGGTTCAGG Forms GGGG Tetrade and G(C-A) Triad. | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*TP*CP*AP*GP*G)-3') | | Authors: | Kettani, A, Basu, G, Gorin, A, Majumdar, A, Skripkin, E, Patel, D.J. | | Deposit date: | 2000-06-06 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A two-stranded template-based approach to G.(C-A) triad formation: designing novel structural elements into an existing DNA framework.
J.Mol.Biol., 301, 2000
|
|
135D
 
 | |
4K97
 
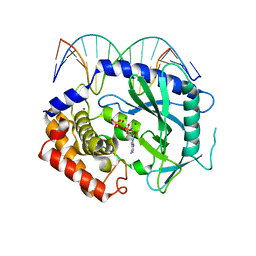 | | Structure of Ternary Complex of cGAS with dsDNA and Bound ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclic GMP-AMP synthase, DNA-F, ... | | Authors: | Gao, P, Wu, Y, Patel, D.J. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase.
Cell(Cambridge,Mass.), 153, 2013
|
|
4K9A
 
 | | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -pG(2 ,5 )pA | | Descriptor: | ADENOSINE MONOPHOSPHATE, Cyclic GMP-AMP synthase, DNA-F, ... | | Authors: | Gao, P, Wu, Y, Patel, D.J. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.264 Å) | | Cite: | Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase.
Cell(Cambridge,Mass.), 153, 2013
|
|
6B46
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF1 | | Descriptor: | Anti-CRISPR protein AcrF1, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy3, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
4K99
 
 | | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -pppdG(2 ,5 )pdG | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, 3'-deoxy-guanosine 5'-monophosphate, Cyclic GMP-AMP synthase, ... | | Authors: | Gao, P, Wu, Y, Patel, D.J. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase.
Cell(Cambridge,Mass.), 153, 2013
|
|
5DDO
 
 | |
1Y26
 
 | | A-riboswitch-adenine complex | | Descriptor: | ADENINE, MAGNESIUM ION, Vibrio vulnificus A-riboswitch | | Authors: | Serganov, A, Yuan, Y.R, Patel, D.J. | | Deposit date: | 2004-11-20 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Discriminative Regulation of Gene Expression by Adenine- and Guanine-Sensing mRNAs
Chem.Biol., 11, 2004
|
|
