6K1E
 
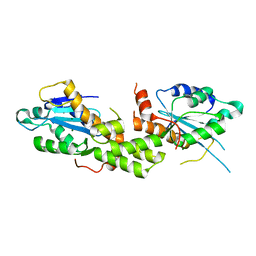 | | Crystal structure of EXD2 exonuclease domain soaked in Mg and GMP | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MAGNESIUM ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
6K1B
 
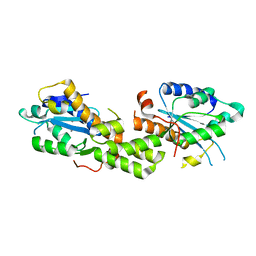 | | Crystal structure of EXD2 exonuclease domain soaked in Mn and dGMP | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MANGANESE (II) ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
7CZ3
 
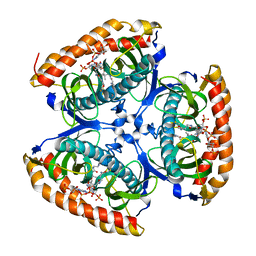 | |
8VJC
 
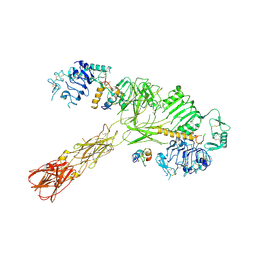 | | Cryo-EM structure of short form insulin receptor (IR-A) with three IGF2 bound, asymmetric conformation. | | Descriptor: | Insulin-like growth factor II, Isoform Short of Insulin receptor | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2024-01-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
8VJB
 
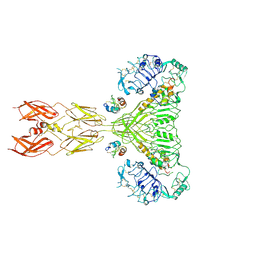 | | Cryo-EM structure of short form insulin receptor (IR-A) with four IGF2 bound, symmetric conformation. | | Descriptor: | Insulin-like growth factor II, Isoform Short of Insulin receptor | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2024-01-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
7YXA
 
 | | XFEL crystal structure of the human sphingosine 1 phosphate receptor 5 in complex with ONO-5430608 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[6-(2-naphthalen-1-ylethoxy)-2,3,4,5-tetrahydro-1H-3-benzazepin-3-ium-3-yl]butanoic acid, ... | | Authors: | Lyapina, E, Marin, E, Gusach, A, Orekhov, P, Gerasimov, A, Luginina, A, Vakhrameev, D, Ergasheva, M, Kovaleva, M, Khusainov, G, Khorn, P, Shevtsov, M, Kovalev, K, Okhrimenko, I, Bukhdruker, S, Popov, P, Hu, H, Weierstall, U, Liu, W, Cho, Y, Gushchin, I, Rogachev, A, Bourenkov, G, Park, S, Park, G, Huyn, H.J, Park, J, Gordeliy, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for receptor selectivity and inverse agonism in S1P 5 receptors.
Nat Commun, 13, 2022
|
|
9AVL
 
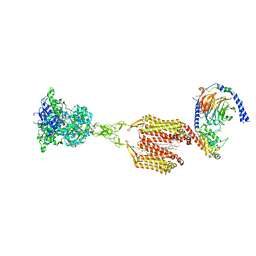 | | Structure of human calcium-sensing receptor in complex with Gi3 protein in nanodiscs | | Descriptor: | (19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-04 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
9AYF
 
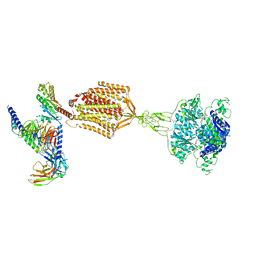 | | Structure of human calcium-sensing receptor in complex with Gi1 (miniGi1) protein in detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-chlorophenyl)-N-[(1R)-1-(3-methoxyphenyl)ethyl]propan-1-amine, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-07 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
9AXF
 
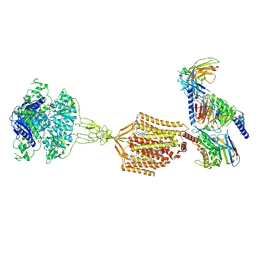 | | Structure of human calcium-sensing receptor in complex with chimeric Gq (miniGisq) protein in detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-chlorophenyl)-N-[(1R)-1-(3-methoxyphenyl)ethyl]propan-1-amine, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
9AVG
 
 | | Structure of human calcium-sensing receptor in complex with chimeric Gs (miniGis) protein in nanodiscs | | Descriptor: | (19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-02 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
9ASB
 
 | | Structure of human calcium-sensing receptor in complex with chimeric Gq (miniGisq) protein in nanodiscs | | Descriptor: | (19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-02-24 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
8GOB
 
 | | Crystal Structure of Glycerol Dehydrogenase in the presence of NAD+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycerol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Park, T, Hoang, H.N, Kang, J.Y, Park, J, Mun, S.A, Jin, M, Yang, J, Jung, C.-H, Eom, S.H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional insights into the flexible beta-hairpin of glycerol dehydrogenase.
Febs J., 290, 2023
|
|
8GOA
 
 | | Crystal Structure of Glycerol Dehydrogenase in the absence of NAD+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycerol dehydrogenase, ZINC ION | | Authors: | Park, T, Hoang, H.N, Kang, J.Y, Park, J, Mun, S.A, Jin, M, Yang, J, Jung, C.-H, Eom, S.H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insights into the flexible beta-hairpin of glycerol dehydrogenase.
Febs J., 290, 2023
|
|
8JQV
 
 | |
2RVC
 
 | | Solution structure of Zalpha domain of goldfish ZBP-containing protein kinase | | Descriptor: | Interferon-inducible and double-stranded-dependent eIF-2kinase | | Authors: | Lee, A, Park, C, Park, J, Kwon, M, Choi, Y, Kim, K, Choi, B, Lee, J. | | Deposit date: | 2015-07-08 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Z-DNA binding domain of PKR-like protein kinase from Carassius auratus and quantitative analyses of the intermediate complex during B-Z transition.
Nucleic Acids Res., 44, 2016
|
|
6XCQ
 
 | | Erythromycin esterase EreC, mutant H289N in its closed conformation | | Descriptor: | EreC, S-1,2-PROPANEDIOL | | Authors: | Zielinski, M, Park, J, Berghuis, A.M. | | Deposit date: | 2020-06-09 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional insights into esterase-mediated macrolide resistance.
Nat Commun, 12, 2021
|
|
4WP0
 
 | | Crystal structure of human kynurenine aminotransferase-I with a C-terminal V5-hexahistidine tag | | Descriptor: | Kynurenine--oxoglutarate transaminase 1 | | Authors: | Nadvi, N.A, Salam, N.K, Park, J, Akladios, F.N, Kapoor, V, Collyer, C.A, Gorrell, M.D, Church, W.B. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human kynurenine aminotransferase-I in a novel space group
To be published
|
|
4WLJ
 
 | | High resolution crystal structure of human kynurenine aminotransferase-I in complex with aminooxyacetate | | Descriptor: | 4'-DEOXY-4'-ACETYLYAMINO-PYRIDOXAL-5'-PHOSPHATE, Kynurenine--oxoglutarate transaminase 1 | | Authors: | Nadvi, N.A, Salam, N.K, Park, J, Akladios, F.N, Kapoor, V, Collyer, C.A, Gorrell, M.D, Church, W.B. | | Deposit date: | 2014-10-07 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | High resolution crystal structures of human kynurenine aminotransferase-I bound to PLP cofactor, and in complex with aminooxyacetate.
Protein Sci., 26, 2017
|
|
4WLH
 
 | | High resolution crystal structure of human kynurenine aminotransferase-I bound to PLP cofactor | | Descriptor: | Kynurenine--oxoglutarate transaminase 1 | | Authors: | Nadvi, N.A, Salam, N.K, Park, J, Akladios, F.N, Kapoor, V, Collyer, C.A, Gorrell, M.D, Church, W.B. | | Deposit date: | 2014-10-07 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | High resolution crystal structures of human kynurenine aminotransferase-I bound to PLP cofactor, and in complex with aminooxyacetate.
Protein Sci., 26, 2017
|
|
2A2K
 
 | | Crystal Structure of an active site mutant, C473S, of Cdc25B Phosphatase Catalytic Domain | | Descriptor: | CHLORIDE ION, M-phase inducer phosphatase 2, SULFATE ION | | Authors: | Sohn, J, Parks, J, Buhrman, G, Brown, P, Kristjansdottir, K, Safi, A, Yang, W, Edelsbrunner, H, Rudolph, J. | | Deposit date: | 2005-06-22 | | Release date: | 2006-01-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Experimental Validation of the Docking Orientation of Cdc25 with Its Cdk2-CycA Protein Substrate.
Biochemistry, 44, 2005
|
|
8IEC
 
 | | Cryo-EM structure of miniGo-scFv16 of GPR156-miniGo-scFv16 complex (local refine) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(o) subunit alpha, ... | | Authors: | Shin, J, Park, J, Cho, Y. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Constitutive activation mechanism of a class C GPCR.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8IEQ
 
 | | Cryo-EM structure of G-protein free GPR156 | | Descriptor: | Probable G-protein coupled receptor 156, [(2R)-3-[(E)-hexadec-9-enoyl]oxy-2-octadecanoyloxy-propyl] 2-(trimethylazaniumyl)ethyl phosphate | | Authors: | Shin, J, Park, J, Cho, Y. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Constitutive activation mechanism of a class C GPCR.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8IEI
 
 | |
8IEB
 
 | |
8IEP
 
 | |
