3OYN
 
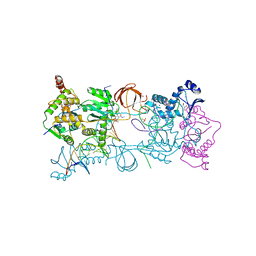 | | Crystal structure of the PFV N224H mutant intasome bound to magnesium and the INSTI MK2048 | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, AMMONIUM ION, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-23 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OYJ
 
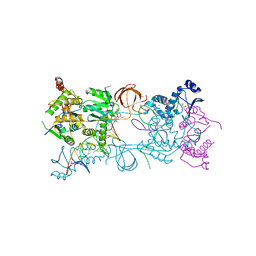 | | Crystal structure of the PFV S217Q mutant intasome in complex with magnesium and the INSTI MK2048 | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, AMMONIUM ION, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-23 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OS0
 
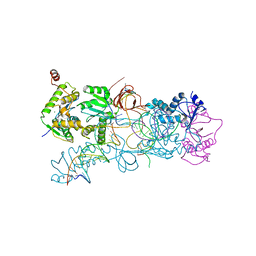 | | PFV strand transfer complex (STC) at 2.81 A resolution | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*CP*CP*CP*GP*AP*G*GP*CP*AP*CP*GP*TP*G)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | Authors: | Maertens, G.N, Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-08 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The mechanism of retroviral integration from X-ray structures of its key intermediates
Nature, 468, 2010
|
|
3OYE
 
 | |
3OYL
 
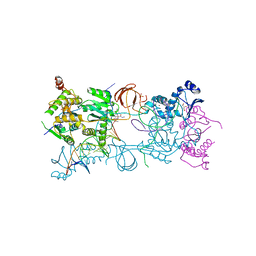 | | Crystal structure of the PFV S217H mutant intasome bound to magnesium and the INSTI MK2048 | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, AMMONIUM ION, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-23 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OYH
 
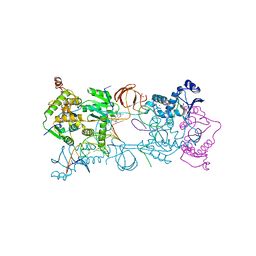 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI MK0536 | | Descriptor: | 6-(3-chloro-4-fluorobenzyl)-4-hydroxy-N,N-dimethyl-2-(1-methylethyl)-3,5-dioxo-2,3,5,6,7,8-hexahydro-2,6-naphthyridine-1-carboxamide, AMMONIUM ION, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-23 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OYA
 
 | |
3OY9
 
 | |
3OYM
 
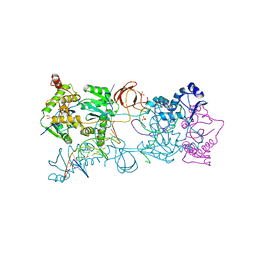 | | Crystal structure of the PFV N224H mutant intasome bound to manganese | | Descriptor: | AMMONIUM ION, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-23 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7Z1Z
 
 | | MVV strand transfer complex (STC) intasome in complex with LEDGF/p75 at 3.5 A resolution | | Descriptor: | DNA (37-MER), DNA (5'-D(*GP*CP*TP*GP*CP*GP*AP*GP*AP*TP*CP*CP*GP*CP*TP*CP*CP*GP*GP*TP*G)-3'), DNA (5'-D(P*TP*TP*GP*AP*TP*TP*AP*GP*GP*GP*TP*G)-3'), ... | | Authors: | Pye, V.E, Ballandras-Colas, A, Cherepanov, P. | | Deposit date: | 2022-02-25 | | Release date: | 2022-05-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Multivalent interactions essential for lentiviral integrase function.
Nat Commun, 13, 2022
|
|
6SP2
 
 | | CryoEM structure of SERINC from Drosophila melanogaster | | Descriptor: | CARDIOLIPIN, Lauryl Maltose Neopentyl Glycol, Membrane protein TMS1d, ... | | Authors: | Pye, V.E, Nans, A, Cherepanov, P. | | Deposit date: | 2019-08-30 | | Release date: | 2020-01-01 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | A bipartite structural organization defines the SERINC family of HIV-1 restriction factors.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6I48
 
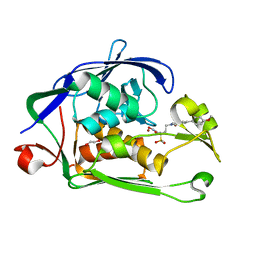 | | Structure of P. aeruginosa LpxC with compound 12: (2R)-4-(6-(2-Fluoro-4-methoxyphenyl)-3-oxo-1H-pyrrolo[1,2-c]imidazol-2(3H)-yl)-N-hydroxy-2-methyl-2-(methylsulfonyl)butanamide | | Descriptor: | (2~{R})-4-[6-(2-fluoranyl-4-methoxy-phenyl)-3-oxidanylidene-1~{H}-pyrrolo[1,2-c]imidazol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, 1,2-ETHANEDIOL, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
1AWW
 
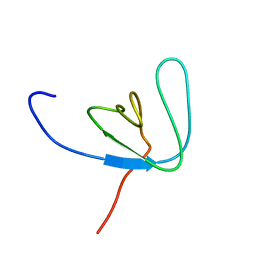 | | SH3 DOMAIN FROM BRUTON'S TYROSINE KINASE, NMR, 42 STRUCTURES | | Descriptor: | BRUTON'S TYROSINE KINASE | | Authors: | Hansson, H, Mattsson, P.T, Allard, P, Haapaniemi, P, Vihinen, M, Smith, C.I.E, Hard, T. | | Deposit date: | 1997-10-06 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain from Bruton's tyrosine kinase.
Biochemistry, 37, 1998
|
|
8POE
 
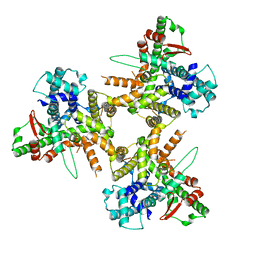 | | Structure of tissue-specific lipid scramblase ATG9B homotrimer, refined with C3 symmetry applied | | Descriptor: | Autophagy-related protein 9B | | Authors: | Chiduza, G.N, Pye, V.E, Tooze, S.A, Cherepanov, P. | | Deposit date: | 2023-07-04 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | ATG9B is a tissue-specific homotrimeric lipid scramblase that can compensate for ATG9A.
Autophagy, 20, 2024
|
|
7ZPP
 
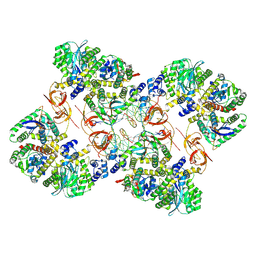 | | Cryo-EM structure of the MVV CSC intasome at 4.5A resolution | | Descriptor: | Integrase, vDNA, non-transferred strand, ... | | Authors: | Ballandras-Colas, A, Maskell, D, Pye, V.E, Locke, J, Swuec, S, Kotecha, A, Costa, A, Cherepanov, P. | | Deposit date: | 2022-04-28 | | Release date: | 2022-05-11 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | A supramolecular assembly mediates lentiviral DNA integration
Science, 355, 2017
|
|
8A1P
 
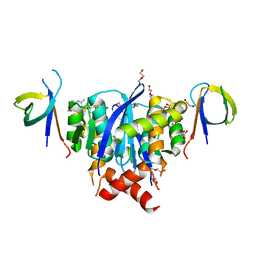 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor BI-D | | Descriptor: | (2S)-tert-butoxy[4-(3,4-dihydro-2H-chromen-6-yl)-2-methylquinolin-3-yl]ethanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, M.R, Pye, V.E, Cook, N.J, Cherepanov, P. | | Deposit date: | 2022-06-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Drug-Induced Interface That Drives HIV-1 Integrase Hypermultimerization and Loss of Function.
Mbio, 14, 2023
|
|
8A1Q
 
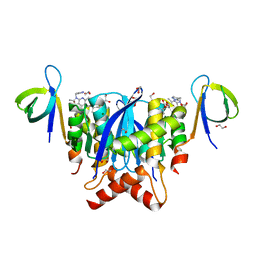 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor STP0404 (Pirmitegravir) | | Descriptor: | (2S)-tert-butoxy{4-(4-chlorophenyl)-2,3,6-trimethyl-1-[(1-methyl-1H-pyrazol-4-yl)methyl]-1H-pyrrolo[2,3-b]pyridin-5-yl}acetic acid, 1,2-ETHANEDIOL, Integrase, ... | | Authors: | Singer, M.R, Pye, V.E, Cook, N.J, Cherepanov, P. | | Deposit date: | 2022-06-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The Drug-Induced Interface That Drives HIV-1 Integrase Hypermultimerization and Loss of Function.
Mbio, 14, 2023
|
|
3OS1
 
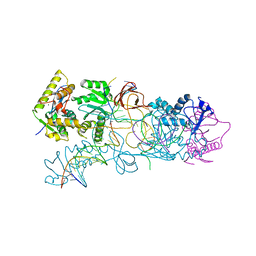 | | PFV target capture complex (TCC) at 2.97 A resolution | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*CP*CP*CP*GP*AP*GP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*CP*TP*CP*GP*GP*G)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*(2DA))-3'), ... | | Authors: | Maertens, G.N, Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-08 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | The mechanism of retroviral integration from X-ray structures of its key intermediates
Nature, 468, 2010
|
|
3OYB
 
 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI MK2048 | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, AMMONIUM ION, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-23 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6I47
 
 | | Structure of P. aeruginosa LpxC with compound 10: (2RS)-4-(5-(2-Fluoro-4-methoxyphenyl)-1-oxoisoindolin-2-yl)-N-hydroxy-2-methyl-2-(methylsulfonyl)butanamide | | Descriptor: | (2~{R})-4-[6-(2-fluoranyl-4-methoxy-phenyl)-3-oxidanylidene-1~{H}-isoindol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, (2~{S})-4-[6-(2-fluoranyl-4-methoxy-phenyl)-3-oxidanylidene-1~{H}-isoindol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
6I49
 
 | | Structure of P. aeruginosa LpxC with compound 17a: (2R)-N-Hydroxy-2-methyl-2-(methylsulfonyl)-4(6((4(morpholinomethyl)phenyl)ethynyl)-3-oxo-1H-pyrrolo[1,2-c]imidazol-2(3H)yl)butanamide | | Descriptor: | (2~{R})-2-methyl-2-methylsulfonyl-4-[6-[2-[4-(morpholin-4-ylmethyl)phenyl]ethynyl]-3-oxidanylidene-1~{H}-pyrrolo[1,2-c]imidazol-2-yl]-~{N}-oxidanyl-butanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
1AWX
 
 | | SH3 DOMAIN FROM BRUTON'S TYROSINE KINASE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BRUTON'S TYROSINE KINASE | | Authors: | Hansson, H, Mattsson, P.T, Allard, P, Haapaniemi, P, Vihinen, M, Smith, C.I.E, Hard, T. | | Deposit date: | 1997-10-06 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain from Bruton's tyrosine kinase.
Biochemistry, 37, 1998
|
|
6I4A
 
 | | Structure of P. aeruginosa LpxC with compound 18d: (2R)-N-Hydroxy-4-(6-((1-(hydroxymethyl)cyclopropyl)buta-1,3-diyn-1-yl)-3-oxo-1H-pyrrolo[1,2-c]imidazol-2(3H)-yl)-2-methyl-2-(methylsulfonyl)butanamide | | Descriptor: | (2~{R})-4-[6-[4-[1-(hydroxymethyl)cyclopropyl]buta-1,3-diynyl]-3-oxidanylidene-1~{H}-pyrrolo[1,2-c]imidazol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
6I46
 
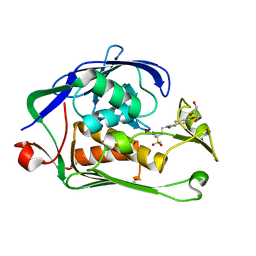 | | Structure of P. aeruginosa LpxC with compound 8: (2RS)-4-(5-(2-Fluoro-4-methoxyphenyl)-2-oxooxazol-3(2H)-yl)-N-hydroxy-2-methyl-2-(methylsulfonyl)butanamide | | Descriptor: | (2~{R})-4-[5-(2-fluoranyl-4-methoxy-phenyl)-2-oxidanylidene-1,3-oxazol-3-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, GLYCEROL, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
3OYD
 
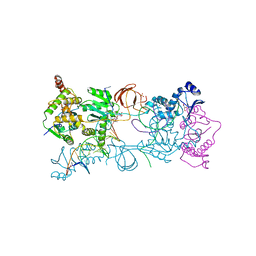 | |
