3LWE
 
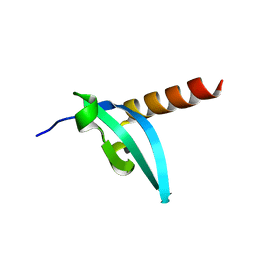 | | The crystal structure of MPP8 | | Descriptor: | M-phase phosphoprotein 8 | | Authors: | Li, Z, Li, Z, Ruan, J, Xu, C, Tong, Y, Pan, P.W, Tempel, W, Crombet, L, Min, J, Zang, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-23 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for specific binding of human MPP8 chromodomain to histone H3 methylated at lysine 9.
Plos One, 6, 2011
|
|
2GWH
 
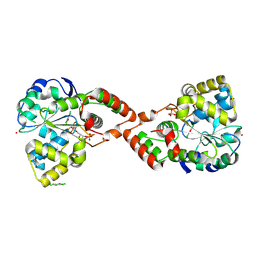 | | Human Sulfotranferase SULT1C2 in complex with PAP and pentachlorophenol | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, PENTACHLOROPHENOL, Sulfotransferase 1C2, ... | | Authors: | Tempel, W, Pan, P.W, Dombrovski, L, Allali-Hassani, A, Vedadi, M, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-04 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and chemical profiling of the human cytosolic sulfotransferases.
Plos Biol., 5, 2007
|
|
3R93
 
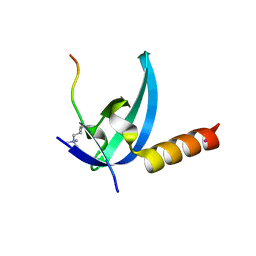 | | Crystal structure of the chromo domain of M-phase phosphoprotein 8 bound to H3K9Me3 peptide | | Descriptor: | H3K9Me3 peptide, M-phase phosphoprotein 8, UNKNOWN ATOM OR ION | | Authors: | Li, J, Li, Z, Ruan, J, Xu, C, Tong, Y, Pan, P.W, Tempel, W, Crombet, L, Min, J, Zang, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-03-24 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.057 Å) | | Cite: | Structural basis for specific binding of human MPP8 chromodomain to histone H3 methylated at lysine 9.
Plos One, 6, 2011
|
|
3CEY
 
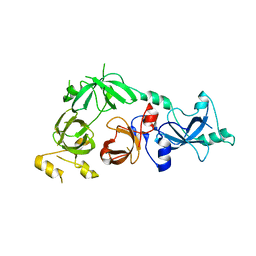 | | Crystal structure of L3MBTL2 | | Descriptor: | Lethal(3)malignant brain tumor-like 2 protein | | Authors: | Nady, N, Guo, Y, Pan, P, Allali-Hassani, A, Qi, C, Zhu, H, Dong, A, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Edwards, A.M, Weigelt, J, Bountra, C, Arrowsmith, C.H, Bochkarev, A, Read, R, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-29 | | Release date: | 2008-05-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2.
Nucleic Acids Res., 37, 2009
|
|
3F70
 
 | | Crystal structure of L3MBTL2-H4K20me1 complex | | Descriptor: | Lethal(3)malignant brain tumor-like 2 protein, N-METHYL-LYSINE | | Authors: | Guo, Y, Qi, C, Allali-Hassani, A, Pan, P, Zhu, H, Dong, A, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Edwards, A.M, Weigelt, J, Bountra, C, Arrowsmith, C.H, Botchkarev, A, Read, R, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-07 | | Release date: | 2009-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2.
Nucleic Acids Res., 37, 2009
|
|
2REO
 
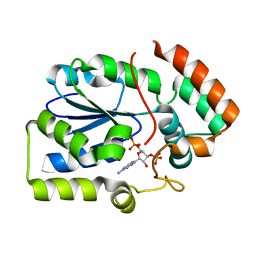 | | Crystal structure of human sulfotransferase 1C3 (Sult1C3) in complex with PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Putative sulfotransferase 1C3 | | Authors: | Tempel, W, Pan, P, Dong, A, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-26 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Crystal structure of human sulfotransferase 1C3 (Sult1C3) in complex with PAP.
To be Published
|
|
3OW8
 
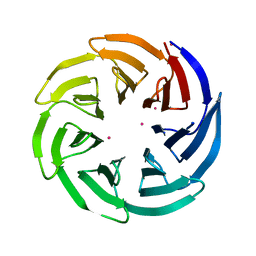 | | Crystal Structure of the WD repeat-containing protein 61 | | Descriptor: | UNKNOWN ATOM OR ION, WD repeat-containing protein 61 | | Authors: | Tempel, W, Li, Z, Chao, X, Lam, R, Wernimont, A.K, He, H, Seitova, A, Pan, P.W, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
4OIM
 
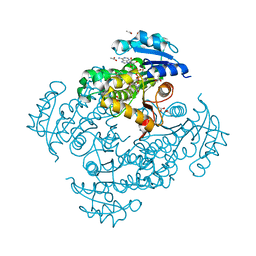 | | Crystal structure of Mycobacterium tuberculosis InhA in complex with inhibitor PT119 in 2.4 M acetate | | Descriptor: | 2-(2-CYANOPHENOXY)-5-HEXYLPHENOL, ACETATE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-01-20 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Time-Dependent Diaryl Ether Inhibitors of InhA: Structure-Activity Relationship Studies of Enzyme Inhibition, Antibacterial Activity, and in vivo Efficacy.
Chemmedchem, 9, 2014
|
|
4OHU
 
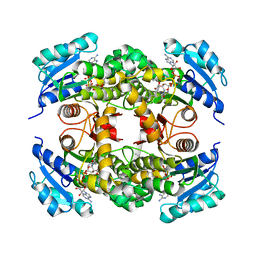 | | Crystal structure of Mycobacterium tuberculosis InhA in complex with inhibitor PT92 | | Descriptor: | 2-(2-bromophenoxy)-5-hexylphenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Yu, W, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-01-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
4OYR
 
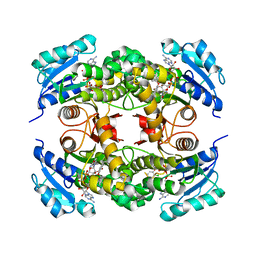 | | Competition of the small inhibitor PT91 with large fatty acyl substrate of the Mycobacterium tuberculosis enoyl-ACP reductase InhA by induced substrate-binding loop refolding | | Descriptor: | 2-(2-chloranylphenoxy)-5-hexyl-phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-13 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2995 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
4OXY
 
 | | Substrate-binding loop movement with inhibitor PT10 in the tetrameric Mycobacterium tuberculosis enoyl-ACP reductase InhA | | Descriptor: | 5-hexyl-2-(2-nitrophenoxy)phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Sullivan, T.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-09 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3501 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
4OXN
 
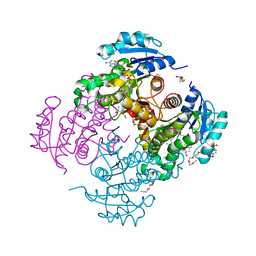 | | Substrate-like binding mode of inhibitor PT155 to the Mycobacterium tuberculosis enoyl-ACP reductase InhA | | Descriptor: | 3,6,9,12,15-pentaoxaoctadecan-17-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-(4-amino-2-methylphenoxy)-2-hexyl-4-hydroxy-1-methylpyridinium, ... | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-05 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2926 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
4OXK
 
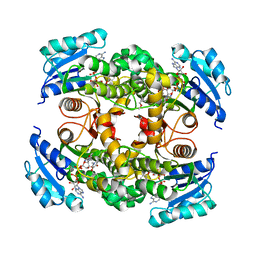 | | Multiple binding modes of inhibitor PT155 to the Mycobacterium tuberculosis enoyl-ACP reductase InhA within a tetramer | | Descriptor: | 3,6,9,12,15-pentaoxaoctadecan-17-amine, 5-(4-amino-2-methylphenoxy)-2-hexyl-4-hydroxy-1-methylpyridinium, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-05 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8429 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
3L2W
 
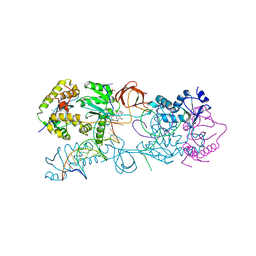 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with manganese and GS9137 (Elvitegravir) | | Descriptor: | 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*TP*GP*TP*A)-3', 5'-D(*TP*AP*CP*AP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3', 6-(3-chloro-2-fluorobenzyl)-1-[(1S)-1-(hydroxymethyl)-2-methylpropyl]-7-methoxy-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, ... | | Authors: | Hare, S, Gupta, S.S, Cherepanov, P. | | Deposit date: | 2009-12-15 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Retroviral intasome assembly and inhibition of DNA strand transfer
Nature, 464, 2010
|
|
5CPF
 
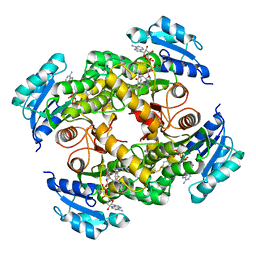 | | Compensation of the effect of isoleucine to alanine mutation by designed inhibition in the InhA enzyme | | Descriptor: | 2-(2-methylphenoxy)-5-[(4-phenyl-1H-1,2,3-triazol-1-yl)methyl]phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.-J, Lai, C.-T, Pan, P, Yu, W, Shah, S, Bommineni, G.R, Perrone, V, Garcia-Diaz, M, Tonge, P.J, Simmerling, C. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.409 Å) | | Cite: | Rational Modulation of the Induced-Fit Conformational Change for Slow-Onset Inhibition in Mycobacterium tuberculosis InhA.
Biochemistry, 54, 2015
|
|
2A1E
 
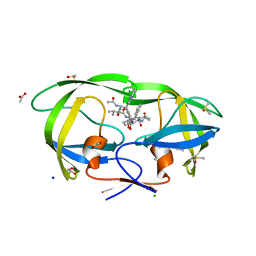 | | High resolution structure of HIV-1 PR with TS-126 | | Descriptor: | ACETATE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Demitri, N, Geremia, S, Randaccio, L, Wuerges, J, Benedetti, F, Berti, F, Dinon, F, Campaner, P, Tell, G. | | Deposit date: | 2005-06-20 | | Release date: | 2006-02-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A potent HIV protease inhibitor identified in an epimeric mixture by high-resolution protein crystallography.
Chemmedchem, 1, 2006
|
|
4N2X
 
 | | Crystal Structure of DL-2-haloacid dehalogenase | | Descriptor: | DL-2-haloacid dehalogenase, GLYCEROL | | Authors: | Siwek, A, Omi, R, Hirotsu, K, Jitsumori, K, Esaki, N, Kurihara, T, Paneth, P. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding modes of DL-2-haloacid dehalogenase revealed by crystallography, modeling and isotope effects studies.
Arch.Biochem.Biophys., 540, 2013
|
|
7PEL
 
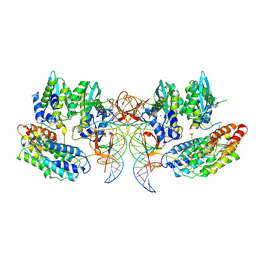 | | CryoEM structure of simian T-cell lymphotropic virus intasome in complex with PP2A regulatory subunit B56 gamma | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), Isoform 3 of PC4 and SFRS1-interacting protein,Isoform Gamma-2 of Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, ... | | Authors: | Barski, M, Pye, V.E, Nans, A, Cherepanov, P, Maertens, G.N. | | Deposit date: | 2021-08-10 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structure of the deltaretroviral intasome in complex with the PP2A regulatory subunit B56gamma.
Nat Commun, 11, 2020
|
|
3BDN
 
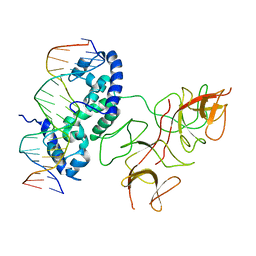 | | Crystal Structure of the Lambda Repressor | | Descriptor: | DNA (5'-D(*DAP*DAP*DTP*DAP*DCP*DCP*DAP*DCP*DTP*DGP*DGP*DCP*DGP*DGP*DTP*DGP*DAP*DTP*DAP*DT)-3'), DNA (5'-D(*DTP*DAP*DTP*DAP*DTP*DCP*DAP*DCP*DCP*DGP*DCP*DCP*DAP*DGP*DTP*DGP*DGP*DTP*DAP*DT)-3'), Lambda Repressor | | Authors: | Stayrook, S.E, Jaru-Ampornpan, P, Hochschild, A, Lewis, M. | | Deposit date: | 2007-11-15 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.909 Å) | | Cite: | Crystal structure of the lambda repressor and a model for pairwise cooperative operator binding
Nature, 452, 2008
|
|
8B7D
 
 | | Luminal domain of TMEM106B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Pye, V.E, Roustan, C, Cherepanov, P. | | Deposit date: | 2022-09-29 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | TMEM106B is a receptor mediating ACE2-independent SARS-CoV-2 cell entry.
Cell, 186, 2023
|
|
6YA7
 
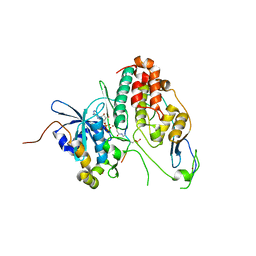 | | Cdc7-Dbf4 bound to an Mcm2-S40 derived bivalent substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division cycle 7-related protein kinase,Cell division cycle 7-related protein kinase,Cell division cycle 7-related protein kinase, DNA replication licensing factor MCM2, ... | | Authors: | Dick, S.D, Cherepanov, P. | | Deposit date: | 2020-03-11 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Basis for the Activation and Target Site Specificity of CDC7 Kinase.
Structure, 28, 2020
|
|
8BUV
 
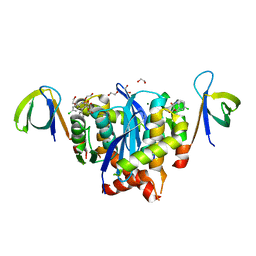 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor LEDGIN 3 | | Descriptor: | (6-chloro-2-oxo-4-phenyl-1,2-dihydroquinolin-3-yl)acetic acid, 1,2-ETHANEDIOL, Integrase, ... | | Authors: | Singer, M.R, Pye, V.E, Cook, N.J, Cherepanov, P. | | Deposit date: | 2022-11-30 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor LEDGIN 3
To Be Published
|
|
8CBR
 
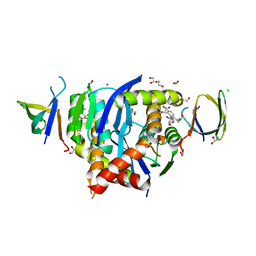 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor BDM-2 | | Descriptor: | (2S)-2-[3-cyclopropyl-2-(3,4-dihydro-2H-chromen-6-yl)-6-methyl-phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8CBU
 
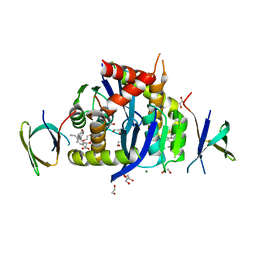 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor MUT884 | | Descriptor: | (2S)-2-[3-cyclopropyl-6-methyl-2-(5-methyl-3,4-dihydro-2H-chromen-6-yl)phenyl]-2-cyclopropyloxy-ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8CBV
 
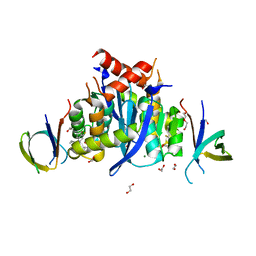 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor MUT916 | | Descriptor: | (2~{S})-2-[3-cyclopropyl-2-(8-fluoranyl-5-methyl-3,4-dihydro-2~{H}-chromen-6-yl)-6-methyl-phenyl]-2-cyclopropyloxy-ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
