2IEG
 
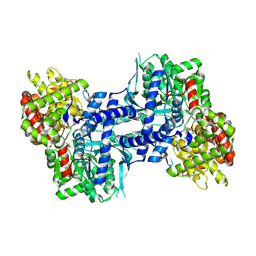 | | Crystal structure of rabbit muscle glycogen phosphorylase in complex with 3,4-dihydro-2-quinolone | | Descriptor: | (2S)-N-[(3S)-1-(2-AMINO-2-OXOETHYL)-2-OXO-1,2,3,4-TETRAHYDROQUINOLIN-3-YL]-2-CHLORO-2H-THIENO[2,3-B]PYRROLE-5-CARBOXAMIDE, (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Glycogen phosphorylase, ... | | Authors: | Birch, A.M, Kenny, P.W, Oikonomakos, N.G, Otterbein, L, Schofield, P, Whittamore, P.R.O, Whalley, D.P, Rowsell, S, Pauptit, R, Pannifer, A, Breed, J, Minshull, C. | | Deposit date: | 2006-09-19 | | Release date: | 2006-12-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of potent, orally active 1-substituted-3,4-dihydro-2-quinolone glycogen phosphorylase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | Descriptor: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
3SNG
 
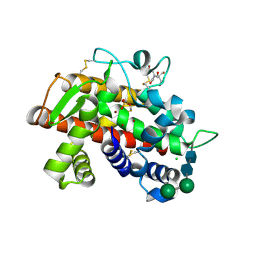 | | X-ray structure of fully glycosylated bifunctional nuclease TBN1 from Solanum lycopersicum (Tomato) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Koval, T, Stepankova, A, Lipovova, P, Podzimek, T, Matousek, J, Duskova, J, Skalova, T, Hasek, J, Dohnalek, J. | | Deposit date: | 2011-06-29 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Plant multifunctional nuclease TBN1 with unexpected phospholipase activity: structural study and reaction-mechanism analysis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GR2
 
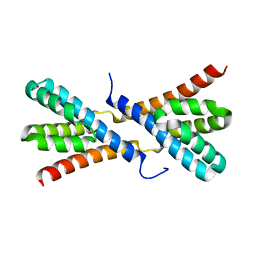 | | Structure of AtRbcX1 from Arabidopsis thaliana. | | Descriptor: | AtRbcX1 | | Authors: | Golik, P, Grudnik, P, Kolesinski, P, Dubin, G, Szczepaniak, A. | | Deposit date: | 2012-08-24 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into eukaryotic Rubisco assembly - Crystal structures of RbcX chaperones from Arabidopsis thaliana.
Biochim.Biophys.Acta, 1830, 2013
|
|
4GR6
 
 | | Crystal structure of AtRbcX2 from Arabidopsis thaliana | | Descriptor: | 1,2-ETHANEDIOL, AtRbcX2 | | Authors: | Grudnik, P, Golik, P, Kolesinski, P, Dubin, G, Szczepaniak, A. | | Deposit date: | 2012-08-24 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into eukaryotic Rubisco assembly - Crystal structures of RbcX chaperones from Arabidopsis thaliana.
Biochim.Biophys.Acta, 1830, 2013
|
|
4Y25
 
 | | Bacterial polysaccharide outer membrane secretin | | Descriptor: | Poly-beta-1,6-N-acetyl-D-glucosamine export protein | | Authors: | Wang, Y, AndolePannuri, A, Ni, D, Zhou, H, Cao, X, Lu, X, Romeo, T, Huang, Y. | | Deposit date: | 2015-02-09 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.821 Å) | | Cite: | Structural Basis for Translocation of a Biofilm-supporting Exopolysaccharide across the Bacterial Outer Membrane
J.Biol.Chem., 291, 2016
|
|
3SK0
 
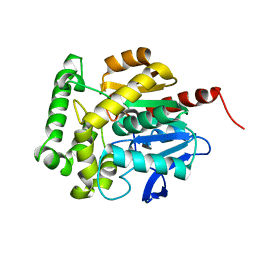 | | structure of Rhodococcus rhodochrous haloalkane dehalogenase DhaA mutant DhaA12 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Koudelakova, T, Damborsky, J, Kuta-Smatanova, I. | | Deposit date: | 2011-06-22 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dynamics and hydration explain failed functional transformation in dehalogenase design.
Nat.Chem.Biol., 10, 2014
|
|
3KW8
 
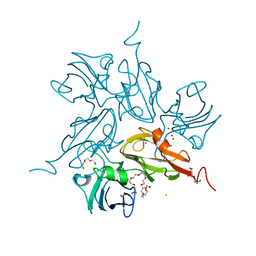 | | Two-domain laccase from Streptomyces coelicolor at 2.3 A resolution | | Descriptor: | COPPER (II) ION, FE (III) ION, Putative copper oxidase, ... | | Authors: | Skalova, T, Dohnalek, J, Kolenko, P, Duskova, J, Stepankova, A, Hasek, J, Ostergaard, L.H, Ostergaard, P.R. | | Deposit date: | 2009-12-01 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of laccase from Streptomyces coelicolor after soaking with potassium hexacyanoferrate and at an improved resolution of 2.3 A
Acta Crystallogr.,Sect.F, 67, 2011
|
|
6B0U
 
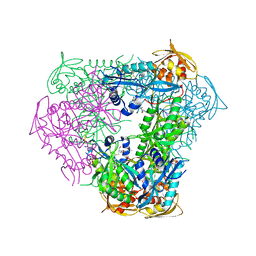 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with a Lys-containing peptide | | Descriptor: | COENZYME A, N-acetyltransferase Eis, Synthetic peptide ATKAPAKKA | | Authors: | Biswas, T, Pang, A.H, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2017-09-15 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Acetylation by Eis and Deacetylation by Rv1151c of Mycobacterium tuberculosis HupB: Biochemical and Structural Insight.
Biochemistry, 57, 2018
|
|
3RK4
 
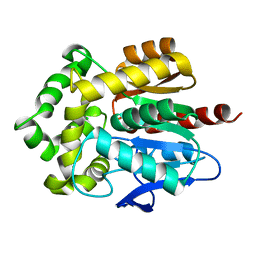 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant DhaA31 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2011-04-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3FBW
 
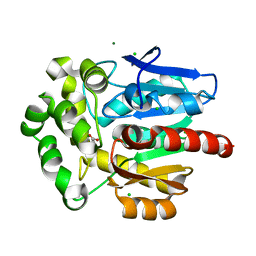 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase DhaA mutant C176Y | | Descriptor: | BENZOIC ACID, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Dohnalek, J, Stsiapanava, A, Gavira, J.A, Kuta Smatanova, I, Kuty, M. | | Deposit date: | 2008-11-20 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Atomic resolution studies of haloalkane dehalogenases DhaA04, DhaA14 and DhaA15 with engineered access tunnels.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3JB4
 
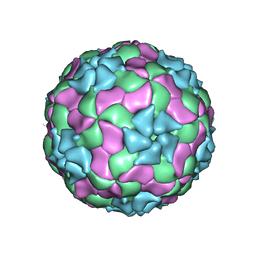 | | Structure of Ljungan virus: insight into picornavirus packaging | | Descriptor: | VP0, VP1, VP3 | | Authors: | Zhu, L, Wang, X.X, Ren, J.S, Porta, C, Wenham, H, Ekstrom, J.-O, Panjwani, A, Knowles, N.J, Kotecha, A, Siebert, A, Lindberg, M, Fry, E.E, Rao, Z.H, Tuthill, T.J, Stuart, D.I. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of Ljungan virus provides insight into genome packaging of this picornavirus.
Nat Commun, 6, 2015
|
|
2YJA
 
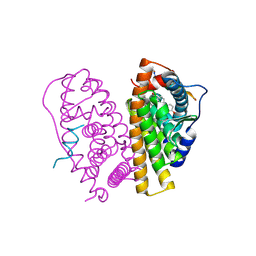 | | Stapled Peptides binding to Estrogen Receptor alpha. | | Descriptor: | ESTRADIOL, ESTROGEN RECEPTOR, STAPLED PEPTIDE | | Authors: | Phillips, C, Roberts, L.R, Schade, M, Bazin, R, Bent, A, Davies, N.L, Irving, S.L, Moore, R, Pannifer, A.D, Brown, D.G, Pickford, A.R, Scott, A, Xu, B. | | Deposit date: | 2011-05-19 | | Release date: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Design and Structure of Stapled Peptides Binding to Estrogen Receptors.
J.Am.Chem.Soc., 133, 2011
|
|
1I5I
 
 | | THE C18S MUTANT OF BOVINE (GAMMA-B)-CRYSTALLIN | | Descriptor: | (GAMMA-B) CRYSTALLIN | | Authors: | Zarutskie, J.A, Asherie, N, Pande, J, Pande, A, Lomakin, J, Lomakin, A, Ogun, O, Stern, L.J, King, J.A, Benedek, G.B. | | Deposit date: | 2001-02-27 | | Release date: | 2001-03-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enhanced crystallization of the Cys18 to Ser mutant of bovine gammaB crystallin.
J.Mol.Biol., 314, 2001
|
|
2YJD
 
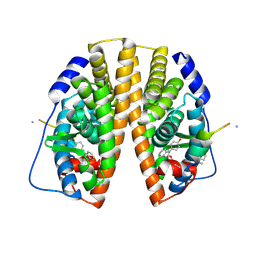 | | Stapled peptide bound to Estrogen Receptor Beta | | Descriptor: | 4-(2-PROPAN-2-YLOXYBENZIMIDAZOL-1-YL)PHENOL, ESTROGEN RECEPTOR BETA, STAPLED PEPTIDE | | Authors: | Phillips, C, Roberts, L.R, Schade, M, Bent, A, Davies, N.L, Moore, R, Pannifer, A.D, Brown, D.G, Pickford, A.R, Irving, S.L. | | Deposit date: | 2011-05-19 | | Release date: | 2011-08-03 | | Last modified: | 2017-01-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design and Structure of Stapled Peptides Binding to Estrogen Receptors.
J.Am.Chem.Soc., 133, 2011
|
|
2YBV
 
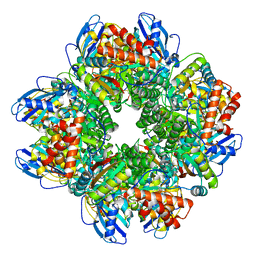 | | STRUCTURE OF RUBISCO FROM THERMOSYNECHOCOCCUS ELONGATUS | | Descriptor: | CHLORIDE ION, RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN, RIBULOSE BISPHOSPHATE CARBOXYLASE SMALL SUBUNIT | | Authors: | Terlecka, B, Wilhelmi, V, Bialek, W, Gubernator, B, Szczepaniak, A, Hofmann, E. | | Deposit date: | 2011-03-10 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Ribulose-1,5-Bisphosphate Carboxylase Oxygenase from Thermosynechococcus Elongatus
To be Published
|
|
2C6D
 
 | | Aurora A kinase activated mutant (T287D) in complex with ADPNP | | Descriptor: | GLYCEROL, PHOSPHATE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Pauptit, R.A, Pannifer, A.D, Breed, J, McMiken, H.H.J, Rowsell, S, Anderson, M. | | Deposit date: | 2005-11-09 | | Release date: | 2006-01-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sar and Inhibitor Complex Structure Determination of a Novel Class of Potent and Specific Aurora Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2BGE
 
 | | Structure-based design of Protein Tyrosine Phosphatase-1B Inhibitors | | Descriptor: | 1,2,5-THIADIAZOLIDIN-3-ONE-1,1-DIOXIDE, PROTEIN-TYROSINE PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Black, E, Breed, J, Breeze, A.L, Embrey, K, Garcia, R, Gero, T.W, Godfrey, L, Kenny, P.W, Morley, A.D, Minshull, C.A, Pannifer, A.D, Read, J, Rees, A, Russell, D.J, Toader, D, Tucker, J. | | Deposit date: | 2004-12-21 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design of Protein Tyrosine Phosphatase-1B Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2C6E
 
 | | Aurora A kinase activated mutant (T287D) in complex with a 5- aminopyrimidinyl quinazoline inhibitor | | Descriptor: | N-{5-[(7-{[(2S)-2-HYDROXY-3-PIPERIDIN-1-YLPROPYL]OXY}-6-METHOXYQUINAZOLIN-4-YL)AMINO]PYRIMIDIN-2-YL}BENZAMIDE, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Pauptit, R.A, Pannifer, A.D, Breed, J, McMiken, H.H.J, Rowsell, S, Anderson, M. | | Deposit date: | 2005-11-09 | | Release date: | 2006-01-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sar and Inhibitor Complex Structure Determination of a Novel Class of Potent and Specific Aurora Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2BGD
 
 | | Structure-based design of Protein Tyrosine Phosphatase-1B Inhibitors | | Descriptor: | 5-(4-METHOXYBIPHENYL-3-YL)-1,2,5-THIADIAZOLIDIN-3-ONE 1,1-DIOXIDE, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Black, E, Breed, J, Breeze, A.L, Embrey, K, Garcia, R, Gero, T.W, Godfrey, L, Kenny, P.W, Morley, A.D, Minshull, C.A, Pannifer, A.D, Read, J, Rees, A, Russell, D.J, Toader, D, Tucker, J. | | Deposit date: | 2004-12-21 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of Protein Tyrosine Phosphatase-1B Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
3FWH
 
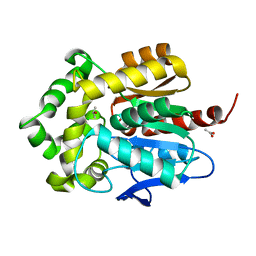 | | Structure of haloalkane dehalogenase mutant Dha15 (I135F/C176Y) from Rhodococcus rhodochrous | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Gavira, J.A, Stsiapanava, A, Kuty, M, Dohnalek, J, Lapkouski, M, Kuta Smatanova, I. | | Deposit date: | 2009-01-18 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Atomic resolution studies of haloalkane dehalogenases DhaA04, DhaA14 and DhaA15 with engineered access tunnels.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3G9X
 
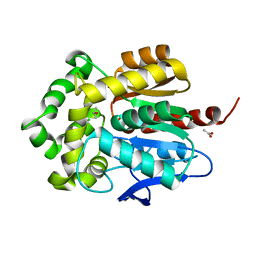 | | Structure of haloalkane dehalogenase DhaA14 mutant I135F from Rhodococcus rhodochrous | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Gavira, J.A, Stsiapanava, A, Kuty, M, Lapkouski, M, Dohnalek, J, Kuta Smatanova, I. | | Deposit date: | 2009-02-15 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Atomic resolution studies of haloalkane dehalogenases DhaA04, DhaA14 and DhaA15 with engineered access tunnels.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3BX4
 
 | | Crystal structure of the snake venom toxin aggretin | | Descriptor: | Aggretin alpha chain, Aggretin beta chain, GLYCEROL, ... | | Authors: | Hooley, E, Papagrigoriou, E, Navdaev, A, Pandey, A, Clemetson, J.M, Clemetson, K.J, Emsley, J. | | Deposit date: | 2008-01-11 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the platelet activator aggretin reveals a novel (alphabeta)2 dimeric structure.
Biochemistry, 47, 2008
|
|
3DR0
 
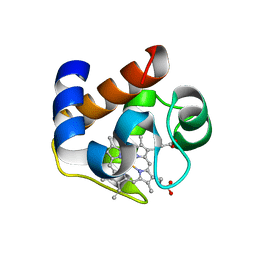 | | Structure of reduced cytochrome c6 from Synechococcus sp. PCC 7002 | | Descriptor: | Cytochrome c6, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Bialek, W, Krzywda, S, Jaskolski, M, Szczepaniak, A. | | Deposit date: | 2008-07-10 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Atomic-resolution structure of reduced cyanobacterial cytochrome c6 with an unusual sequence insertion
Febs J., 276, 2009
|
|
4XJX
 
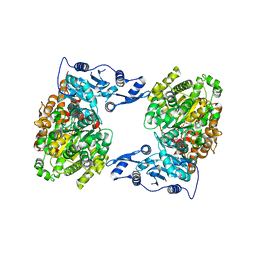 | | STRUCTURE OF MUTANT (E165H) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HsdR, MAGNESIUM ION | | Authors: | Baikova, T, Stsiapanava, A, Moche, M, Degtjarik, O, Kuta-Smatanova, I, Ettrich, R. | | Deposit date: | 2015-01-09 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | STRUCTURE OF MUTANT (E165H) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP
To Be Published
|
|
