2OC7
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH571696 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, TERT-BUTYL {(1S)-2-[(1R,2S,5R)-2-({[(1S)-3-AMINO-1-(CYCLOBUTYLMETHYL)-2,3-DIOXOPROPYL]AMINO}CARBONYL)-7,7-DIMETHYL-6-OXA-3-AZABICYCLO[3.2.0]HEPT-3-YL]-1-CYCLOHEXYL-2-OXOETHYL}CARBAMATE, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OBQ
 
 | | Discovery of the HCV NS3/4A Protease Inhibitor SCH503034. Key Steps in Structure-Based Optimization | | Descriptor: | Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC0
 
 | | Structure of NS3 complexed with a ketoamide inhibitor SCh491762 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, Hepatitis C virus, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OBO
 
 | | Structure of HEPATITIS C VIRAL NS3 protease domain complexed with NS4A peptide and ketoamide SCH476776 | | Descriptor: | BETA-MERCAPTOETHANOL, HCV NS3 protease, HCV NS4A peptide, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC8
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH503034 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C virus, ZINC ION, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R.S, Arasappan, A, Bennett, F, Bogen, S.L, Chen, K, Jao, E, Liu, Y.T, Lovey, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC1
 
 | | Structure of the HCV NS3/4A Protease Inhibitor CVS4819 | | Descriptor: | (2S)-({N-[(3S)-3-({N-[(2S,4E)-2-ISOPROPYL-7-METHYLOCT-4-ENOYL]-L-LEUCYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETI C ACID, Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
7E4J
 
 | | X-ray crystal structure of VapB12 antitoxin from mycobacterium tuberculosis in space group P41. | | Descriptor: | Antitoxin, ZINC ION | | Authors: | Pratap, S, Megta, A.K, Talwar, S, Chandresh, S, Pandey, A.K, Krishnan, V. | | Deposit date: | 2021-02-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray crystal structure of VapB12 antitoxin from mycobacterium tuberculosis in space group P41.
To be published
|
|
3ZW3
 
 | | Fragment based discovery of a novel and selective PI3 Kinase inhibitor | | Descriptor: | N-{6-[(Z)-(2,4-dioxo-1,3-thiazolidin-5-ylidene)methyl]imidazo[1,2-a]pyridin-2-yl}acetamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Brown, D.G, Hughes, S.J, Milan, D.S, Kilty, I.C, Lewthwaite, R.A, Mathias, J.P, O'Reilly, M.A, Pannifer, A, Phelan, A, Baldock, D.A. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fragment based discovery of a novel and selective PI3 kinase inhibitor.
Bioorg. Med. Chem. Lett., 21, 2011
|
|
3VW7
 
 | | Crystal structure of human protease-activated receptor 1 (PAR1) bound with antagonist vorapaxar at 2.2 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Proteinase-activated receptor 1, ... | | Authors: | Zhang, C, Srinivasan, Y, Arlow, D.H, Fung, J.J, Palmer, D, Zheng, Y, Green, H.F, Pandey, A, Dror, R.O, Shaw, D.E, Weis, W.I, Coughlin, S.R, Kobilka, B.K. | | Deposit date: | 2012-08-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution crystal structure of human protease-activated receptor 1
Nature, 492, 2012
|
|
3ZNP
 
 | | IN VITRO AND IN VIVO INHIBITION OF HUMAN D-AMINO ACID OXIDASE: REGULATION OF D-SERINE CONCENTRATION IN THE BRAIN | | Descriptor: | 3-HYDROXY-2H-CHROMEN-2-ONE, D-AMINO-ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hopkins, S.C, Heffernan, M.L.R, Saraswat, L.D, Bowen, C.A, Melnick, L, Hardy, L.W, Orsini, M.A, Allen, M.S, Koch, P, Spear, K.L, Foglesong, R.J, Soukri, M, Chytil, M, Fang, Q.K, Jones, S.W, Varney, M.A, Panatier, A, Oliet, S.H.R, Pollegioni, L, Piubelli, L, Molla, G, Nardini, M, Large, T.H. | | Deposit date: | 2013-02-15 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural, Kinetic, and Pharmacodynamic Mechanisms of D-Amino Acid Oxidase Inhibition by Small Molecules.
J.Med.Chem., 56, 2013
|
|
3ZNO
 
 | | IN VITRO AND IN VIVO INHIBITION OF HUMAN D-AMINO ACID OXIDASE: REGULATION OF D-SERINE CONCENTRATION IN THE BRAIN | | Descriptor: | 4-(4-chlorophenethyl)-1H-pyrrole-2-carboxylic acid, D-AMINO-ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hopkins, S.C, Heffernan, M.L.R, Saraswat, L.D, Bowen, C.A, Melnick, L, Hardy, L.W, Orsini, M.A, Allen, M.S, Koch, P, Spear, K.L, Foglesong, R.J, Soukri, M, Chytil, M, Fang, Q.K, Jones, S.W, Varney, M.A, Panatier, A, Oliet, S.H.R, Pollegioni, L, Piubelli, L, Molla, G, Nardini, M, Large, T.H. | | Deposit date: | 2013-02-15 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural, Kinetic, and Pharmacodynamic Mechanisms of D-Amino Acid Oxidase Inhibition by Small Molecules.
J.Med.Chem., 56, 2013
|
|
3ZNN
 
 | | IN VITRO AND IN VIVO INHIBITION OF HUMAN D-AMINO ACID OXIDASE: REGULATION OF D-SERINE CONCENTRATION IN THE BRAIN | | Descriptor: | 4H-THIENO[3,2-B]PYROLE-5-CARBOXYLIC ACID, D-AMINO-ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hopkins, S.C, Heffernan, M.L.R, Saraswat, L.D, Bowen, C.A, Melnick, L, Hardy, L.W, Orsini, M.A, Allen, M.S, Koch, P, Spear, K.L, Foglesong, R.J, Soukri, M, Chytil, M, Fang, Q.K, Jones, S.W, Varney, M.A, Panatier, A, Oliet, S.H.R, Pollegioni, L, Piubelli, L, Molla, G, Nardini, M, Large, T.H. | | Deposit date: | 2013-02-15 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural, Kinetic, and Pharmacodynamic Mechanisms of D-Amino Acid Oxidase Inhibition by Small Molecules.
J.Med.Chem., 56, 2013
|
|
3ZNQ
 
 | | IN VITRO AND IN VIVO INHIBITION OF HUMAN D-AMINO ACID OXIDASE: REGULATION OF D-SERINE CONCENTRATION IN THE BRAIN | | Descriptor: | 3-PHENETHYL-4H-FURO[3,2-B]PYRROLE-5-CARBOXYLIC ACID, D-AMINO-ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hopkins, S.C, Heffernan, M.L.R, Saraswat, L.D, Bowen, C.A, Melnick, L, Hardy, L.W, Orsini, M.A, Allen, M.S, Koch, P, Spear, K.L, Foglesong, R.J, Soukri, M, Chytil, M, Fang, Q.K, Jones, S.W, Varney, M.A, Panatier, A, Oliet, S.H.R, Pollegioni, L, Piubelli, L, Molla, G, Nardini, M, Large, T.H. | | Deposit date: | 2013-02-15 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural, Kinetic, and Pharmacodynamic Mechanisms of D-Amino Acid Oxidase Inhibition by Small Molecules.
J.Med.Chem., 56, 2013
|
|
3ZXW
 
 | | STRUCTURE OF ACTIVATED RUBISCO FROM THERMOSYNECHOCOCCUS ELONGATUS COMPLEXED WITH 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Terlecka, B, Wilhelmi, V, Bialek, W, Gubernator, B, Szczepaniak, A, Hofmann, E. | | Deposit date: | 2011-08-16 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Ribulose-1,5-Bisphosphate Carboxylase Oxygenase from Thermosynechococcus Elongatus
To be Published
|
|
3ZCW
 
 | | Eg5 - New allosteric binding site | | Descriptor: | (2E)-2-(3-fluoranyl-4-methoxy-phenyl)imino-1-[[2-(trifluoromethyl)phenyl]methyl]-3H-benzimidazole-5-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ulaganathan, V, Talapatra, S.K, Kozielski, F, Pannifer, A.D. | | Deposit date: | 2012-11-23 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.691 Å) | | Cite: | Structural Insights Into a Unique Inhibitor Binding Pocket in Kinesin Spindle Protein.
J.Am.Chem.Soc., 135, 2013
|
|
4EIE
 
 | | Crystal structure of cytochrome c6C from Synechococcus sp. PCC 7002 | | Descriptor: | CHLORIDE ION, Cytochrome c6, HEME C, ... | | Authors: | Krzywda, S, Bialek, W, Zatwarnicki, P, Jaskolski, M, Szczepaniak, A. | | Deposit date: | 2012-04-05 | | Release date: | 2013-04-10 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Cytochrome c6 and c6C from Synechococcus sp. PCC 7002 - structure and function.
To be Published
|
|
4EIC
 
 | |
4EIF
 
 | | Crystal structure of cytochrome c6C L50Q mutant from Synechococcus sp. PCC 7002 | | Descriptor: | CHLORIDE ION, Cytochrome c6, HEME C, ... | | Authors: | Krzywda, S, Bialek, W, Zatwarnicki, P, Jaskolski, M, Szczepaniak, A. | | Deposit date: | 2012-04-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Cytochrome c6 and c6C from Synechococcus sp. PCC 7002 - structure and function.
To be Published
|
|
4DJ4
 
 | | X-ray structure of mutant N211D of bifunctional nuclease TBN1 from Solanum lycopersicum (Tomato) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Nuclease, ... | | Authors: | Koval, T, Stepankova, A, Lipovova, P, Podzimek, T, Matousek, J, Duskova, J, Skalova, T, Hasek, J, Dohnalek, J. | | Deposit date: | 2012-02-01 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Plant multifunctional nuclease TBN1 with unexpected phospholipase activity: structural study and reaction-mechanism analysis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BIQ
 
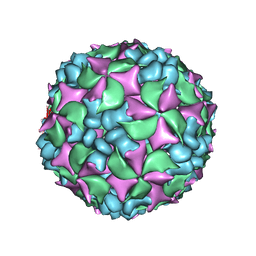 | | Homology model of coxsackievirus A7 (CAV7) empty capsid proteins. | | Descriptor: | VP1, VP2, VP3 | | Authors: | Seitsonen, J.J.T, Shakeel, S, Susi, P, Pandurangan, A.P, Sinkovits, R.S, Hyvonen, H, Laurinmaki, P, Yla-Pelto, J, Topf, M, Hyypia, T, Butcher, S.J. | | Deposit date: | 2013-04-12 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.09 Å) | | Cite: | Combined Approaches to Flexible Fitting and Assessment in Virus Capsids Undergoing Conformational Change.
J.Struct.Biol., 185, 2014
|
|
4BIP
 
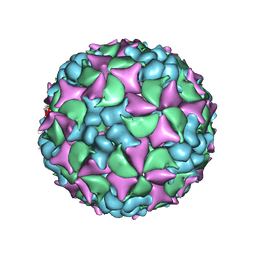 | | Homology model of coxsackievirus A7 (CAV7) full capsid proteins. | | Descriptor: | VP1, VP2, VP3 | | Authors: | Seitsonen, J.J.T, Shakeel, S, Susi, P, Pandurangan, A.P, Sinkovits, R.S, Hyvonen, H, Laurinmaki, P, Yla-Pelto, J, Topf, M, Hyypia, T, Butcher, S.J. | | Deposit date: | 2013-04-12 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.23 Å) | | Cite: | Combined Approaches to Flexible Fitting and Assessment in Virus Capsids Undergoing Conformational Change.
J.Struct.Biol., 185, 2014
|
|
4D1S
 
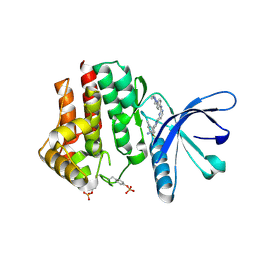 | | Pyrrole-3-carboxamides as potent and selective JAK2 inhibitors | | Descriptor: | 2-(5-chloro-2-methylphenyl)-1-methyl-5-(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)-1H-pyrrole-3-carboxamide, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Bertrand, J, Canevari, G, Fasolini, M, Brasca, M.G, Nesi, M, Avanzi, N, Ballinari, D, Bandiera, T, Bindi, S, Carenzi, D, Casero, D, Ceriani, L, Ciomei, M, Cirla, A, Colombo, M, Cribioli, S, Cristiani, C, Della Vedova, F, Fachin, G, Felder, E.R, Galvani, A, Isacchi, A, Mirizzi, D, Motto, I, Panzeri, A, Pesenti, E, Vianello, P, Gnocchi, P, Donati, D. | | Deposit date: | 2014-05-05 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Pyrrole-3-Carboxamides as Potent and Selective Jak2 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
4D0X
 
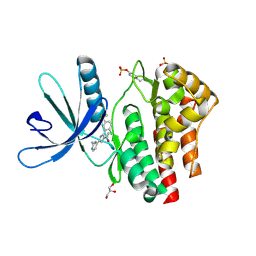 | | Pyrrole-3-carboxamides as potent and selective JAK2 inhibitors | | Descriptor: | 5-(2-aminopyrimidin-4-yl)-2-[2-chloro-5-(trifluoromethyl)phenyl]-1H-pyrrole-3-carboxamide, GLYCEROL, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Canevari, G, Fasolini, M, Bertrand, J, Brasca, M.G, Nesi, M, Avanzi, N, Ballinari, D, Bandiera, T, Bindi, S, Carenzi, D, Casero, D, Ceriani, L, Ciomei, M, Cirla, A, Colombo, M, Cribioli, S, Cristiani, C, Della Vedova, F, Fachin, G, Felder, E.R, Galvani, A, Isacchi, A, Mirizzi, D, Motto, I, Panzeri, A, Pesenti, E, Vianello, P, Gnocchi, P, Donati, D. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Pyrrole-3-Carboxamides as Potent and Selective Jak2 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
4BEX
 
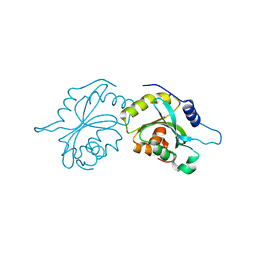 | | Structure of human Cofilin1 | | Descriptor: | COFILIN-1 | | Authors: | Klejnot, M, Gabrielsen, M, Cameron, J, Mleczak, A, Talapatra, S.K, Kozielski, F, Pannifer, A, Olson, M.F. | | Deposit date: | 2013-03-12 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the Human Cofilin1 Structure Reveals Conformational Changes Required for Actin-Binding
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BOM
 
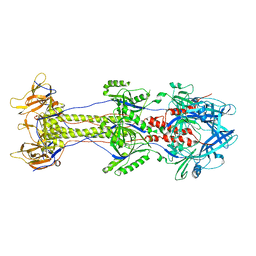 | | Structure of herpesvirus fusion glycoprotein B-bilayer complex revealing the protein-membrane and lateral protein-protein interaction | | Descriptor: | ENVELOPE GLYCOPROTEIN B | | Authors: | Maurer, U.E, Zeev-Ben-Mordehai, Z, Pandurangan, A.P, Cairns, T.M, Hannah, B.P, Whitbeck, J.C, Eisenberg, R.J, Cohen, G.H, Topf, M, Huiskonen, J.T, Grunewald, K. | | Deposit date: | 2013-05-21 | | Release date: | 2013-07-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (27 Å) | | Cite: | The structure of herpesvirus fusion glycoprotein B-bilayer complex reveals the protein-membrane and lateral protein-protein interaction.
Structure, 21, 2013
|
|
