2RF3
 
 | | Crystal Structure of Tricyclo-DNA: An Unusual Compensatory Change of Two Adjacent Backbone Torsion Angles | | Descriptor: | 5'-d(CGCG(TCY)ATTCGCG)-3', SPERMINE, ZINC ION | | Authors: | Pallan, P.S, Ittig, D, Heroux, A, Wawrzak, Z, Leumann, C.J, Egli, M. | | Deposit date: | 2007-09-27 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of tricyclo-DNA: an unusual compensatory change of two adjacent backbone torsion angles.
Chem.Commun.(Camb.), 2008, 2008
|
|
3CJZ
 
 | | Effects of N2,N2-dimethylguanosine on RNA structure and stability: crystal structure of an RNA duplex with tandem m22G:A pairs | | Descriptor: | RNA (5'-R(*GP*GP*AP*CP*GP*(M2G)P*AP*CP*GP*UP*CP*CP*U)-3') | | Authors: | Pallan, P.S, Kreutz, C, Bosio, S, Micura, R, Egli, M. | | Deposit date: | 2008-03-14 | | Release date: | 2008-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effects of N2,N2-dimethylguanosine on RNA structure and stability: crystal structure of an RNA duplex with tandem m2 2G:A pairs.
Rna, 14, 2008
|
|
6XHC
 
 | | Structure of glycinyl 5'-O-adenosine phosphoramidate | | Descriptor: | 2-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]amino]ethanoic acid, Ribonuclease pancreatic | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5W20
 
 | | Crystal Structure of inosine-substituted duplex DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*IP*IP*CP*CP*TP*GP*G)-3'), MAGNESIUM ION | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2017-06-05 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Comparative analysis of inosine-substituted duplex DNA by circular dichroism and X-ray crystallography.
J. Biomol. Struct. Dyn., 36, 2018
|
|
5W1Z
 
 | | Crystal Structure of inosine-substituted decamer duplex DNA (I4) | | Descriptor: | DNA (5'-D(*CP*CP*AP*IP*IP*CP*CP*(BRU)P*IP*I)-3'), MAGNESIUM ION, SODIUM ION | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2017-06-05 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Comparative analysis of inosine-substituted duplex DNA by circular dichroism and X-ray crystallography.
J. Biomol. Struct. Dyn., 36, 2018
|
|
6XHE
 
 | | Structure of beta-prolinyl 5'-O-adenosine phosphoramidate | | Descriptor: | 5'-O-[(S)-[(3S)-3-carboxypyrrolidin-1-yl](hydroxy)phosphoryl]adenosine, ADENOSINE MONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XHF
 
 | | Structure of Phenylalanyl-5'-O-adenosine phosphoramidate | | Descriptor: | (2~{S})-2-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]amino]-3-phenyl-propanoic acid, CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XHD
 
 | | Structure of Prolinyl-5'-O-adenosine phosphoramidate | | Descriptor: | (2~{S})-1-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]pyrrolidine-2-carboxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The Enzyme-Free Release of Nucleotides from Phosphoramidates Depends Strongly on the Amino Acid.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7JJE
 
 | |
7JJD
 
 | |
1PWF
 
 | | One Sugar Pucker Fits All: Pairing Versatility Despite Conformational Uniformity in TNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(2MU)P*(FA2)P*CP*GP*C)-3', SPERMINE | | Authors: | Pallan, P.S, Wilds, C.J, Wawrzak, Z, Krishnamurthy, R, Eschenmoser, A, Egli, M. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Why does TNA cross-pair more strongly with RNA than with DNA? an answer from X-ray analysis.
ANGEW.CHEM.INT.ED.ENGL., 42, 2003
|
|
4Y8W
 
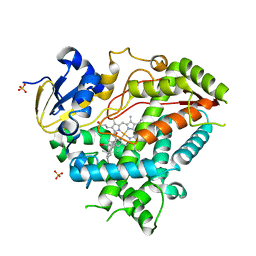 | | Crystal Structure of Human Cytochrome P450 21A2 Progesterone Complex | | Descriptor: | Cytochrome P450 21-hydroxylase, PROGESTERONE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pallan, P.S, Lei, L, Egli, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Human Cytochrome P450 21A2, the Major Steroid 21-Hydroxylase: STRUCTURE OF THE ENZYMEPROGESTERONE SUBSTRATE COMPLEX AND RATE-LIMITING C-H BOND CLEAVAGE.
J.Biol.Chem., 290, 2015
|
|
3D0P
 
 | |
2GUN
 
 | |
8SV4
 
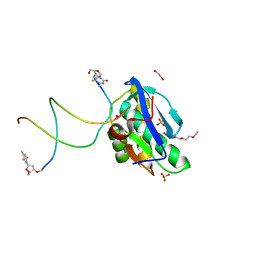 | | 7-Deazapurines and 5-Halogenpyrimidine DNA duplex | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-ethoxyethoxy)ethoxy]ethanol, ACETATE ION, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Morphing by a DNA Analogue Featuring 7-Deazapurines and 5-Halogenpyrimidines and the Origins of Adenine-Tract Geometry.
Biochemistry, 62, 2023
|
|
8SV3
 
 | | 7-Deazapurines and 5-Halogenpyrimidine DNA duplex | | Descriptor: | GLYCEROL, MAGNESIUM ION, Modified DNA, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Conformational Morphing by a DNA Analogue Featuring 7-Deazapurines and 5-Halogenpyrimidines and the Origins of Adenine-Tract Geometry.
Biochemistry, 62, 2023
|
|
4F2Y
 
 | | Structure of 3'-Fluoro Cyclohexenyl Nucleic Acid Decamer | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(XTF)P*AP*CP*GP*C)-3', MAGNESIUM ION | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Synthesis and Antisense Properties of Fluoro Cyclohexenyl Nucleic Acid (F-CeNA), a Nuclease Stable Mimic of 2'-Fluoro RNA.
J.Org.Chem., 77, 2012
|
|
4F2X
 
 | | Structure of 3'-Fluoro Cyclohexenyl Nucleic Acid Heptamer | | Descriptor: | 5'-D(*CP*GP*CP*AP*CP*GP*C)-3', 5'-D(*GP*CP*GP*(XTF)P*GP*CP*G)-3', COBALT (III) ION, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Synthesis and Antisense Properties of Fluoro Cyclohexenyl Nucleic Acid (F-CeNA), a Nuclease Stable Mimic of 2'-Fluoro RNA.
J.Org.Chem., 77, 2012
|
|
5DO4
 
 | | Thrombin-RNA aptamer complex | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2015-09-10 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Evoking picomolar binding in RNA by a single phosphorodithioate linkage.
Nucleic Acids Res., 44, 2016
|
|
5DO5
 
 | |
7U38
 
 | | Pixantrone tethered DNA duplex | | Descriptor: | 1,2-ETHANEDIOL, Pixantrone AP conjugate-modified DNA | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2022-02-26 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural Characterization of a Covalent Conjugate between Pixantrone and an Abasic Site in DNA
To Be Published
|
|
5VBU
 
 | |
5VAJ
 
 | | BhRNase H - amide-RNA/DNA complex | | Descriptor: | ACETATE ION, DNA (5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3'), GLYCEROL, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2017-03-27 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Amide linkages mimic phosphates in RNA interactions with proteins and are well tolerated in the guide strand of short interfering RNAs.
Nucleic Acids Res., 45, 2017
|
|
5V1K
 
 | | Structure of R-GNA dodecamer | | Descriptor: | RNA (5'-R(*CP*GP*CP*GP*AP*AP*(5BU)P*(8RJ)P*AP*GP*CP*G)-3') | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2017-03-02 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Chirality Dependent Potency Enhancement and Structural Impact of Glycol Nucleic Acid Modification on siRNA.
J. Am. Chem. Soc., 2017
|
|
5V1L
 
 | | Structure of S-GNA dodecamer | | Descriptor: | RNA (5'-R(*CP*GP*CP*GP*AP*AP*UP*(ZTH)P*AP*GP*CP*G)-3'), SPERMINE, STRONTIUM ION | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2017-03-02 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Chirality Dependent Potency Enhancement and Structural Impact of Glycol Nucleic Acid Modification on siRNA.
J. Am. Chem. Soc., 139, 2017
|
|
