5TO5
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-10-16 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5TO6
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Bandyopadhyay, A, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-10-16 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5TO7
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-10-16 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5TVB
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-11-08 | | Release date: | 2017-09-20 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
3N95
 
 | |
3N96
 
 | |
3N93
 
 | | Crystal structure of human CRFR2 alpha extracellular domain in complex with Urocortin 3 | | Descriptor: | GLYCEROL, Maltose binding protein-CRFR2 alpha, Urocortin-3, ... | | Authors: | Pal, K, Swaminathan, K, Pioszak, A.A, Xu, H.E. | | Deposit date: | 2010-05-28 | | Release date: | 2010-10-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of ligand selectivity in human CRFR1 and CRFR2 alpha extracellular domain
To be Published
|
|
3K1D
 
 | | Crystal structure of glycogen branching enzyme synonym: 1,4-alpha-D-glucan:1,4-alpha-D-GLUCAN 6-glucosyl-transferase from mycobacterium tuberculosis H37RV | | Descriptor: | 1,4-alpha-glucan-branching enzyme | | Authors: | Pal, K, Kumar, S, Swaminathan, K. | | Deposit date: | 2009-09-27 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of full-length Mycobacterium tuberculosis H37Rv glycogen branching enzyme: insights of N-terminal beta-sandwich in substrate specificity and enzymatic activity
J.Biol.Chem., 285, 2010
|
|
6JIE
 
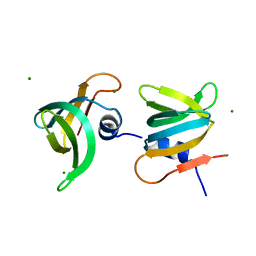 | | YaeO bound to Magnesium from Vibrio cholerae O395 | | Descriptor: | MAGNESIUM ION, YaeO | | Authors: | Pal, K, Yadav, M, Sen, U. | | Deposit date: | 2019-02-20 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Vibrio cholerae YaeO is a Structural Homologue of RNA Chaperone Hfq that Inhibits Rho-dependent Transcription Termination by Dissociating its Hexameric State.
J.Mol.Biol., 431, 2019
|
|
3KJX
 
 | |
2QVC
 
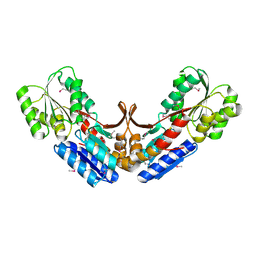 | | Crystal structure of a periplasmic sugar ABC transporter from Thermotoga maritima | | Descriptor: | Sugar ABC transporter, periplasmic sugar-binding protein, beta-D-glucopyranose | | Authors: | Palani, K, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a periplasmic glucose-binding protein from Thermotoga maritima.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3LOP
 
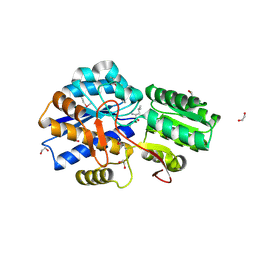 | | Crystal structure of substrate-binding periplasmic protein (Pbp) from Ralstonia solanacearum | | Descriptor: | 1,2-ETHANEDIOL, LEUCINE, MAGNESIUM ION, ... | | Authors: | Palani, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-04 | | Release date: | 2010-02-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of substrate-binding periplasmic protein (Pbp) from Ralstonia solanacearum
To be Published
|
|
3BCV
 
 | |
3BZW
 
 | | Crystal structure of a putative lipase from Bacteroides thetaiotaomicron | | Descriptor: | ACETATE ION, Putative lipase, SULFATE ION | | Authors: | Palani, K, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-18 | | Release date: | 2008-02-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of a putative lipase from Bacteroides thetaiotaomicron.
To be Published
|
|
3KZG
 
 | |
2QXY
 
 | |
3LHL
 
 | | Crystal structure of a putative agmatinase from Clostridium difficile | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Palani, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative agmatinase from Clostridium difficile
To be Published
|
|
3LTO
 
 | |
3N53
 
 | |
3NHM
 
 | |
3MIZ
 
 | |
3SMD
 
 | |
3CO8
 
 | | Crystal structure of alanine racemase from Oenococcus oeni | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Palani, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of alanine racemase from Oenococcus oeni with bound pyridoxal 5'-phosphate.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3EXQ
 
 | |
3FCM
 
 | |
