3H7J
 
 | | Crystal structure of BacB, an enzyme involved in Bacilysin synthesis, in monoclinic form | | Descriptor: | 3-PHENYLPYRUVIC ACID, Bacilysin biosynthesis protein bacB, COBALT (II) ION, ... | | Authors: | Rajavel, M, Gopal, B. | | Deposit date: | 2009-04-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Role of Bacillus subtilis BacB in the synthesis of bacilysin
J.Biol.Chem., 284, 2009
|
|
2O7G
 
 | |
3H9A
 
 | | Crystal structure of BacB, an enzyme involved in Bacilysin synthesis, in triclinic form | | Descriptor: | 3-PHENYLPYRUVIC ACID, Bacilysin biosynthesis protein bacB, COBALT (II) ION, ... | | Authors: | Rajavel, M, Gopal, B. | | Deposit date: | 2009-04-30 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Analysis of multiple crystal forms of Bacillus subtilis BacB suggests a role for a metal ion as a nucleant for crystallization
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3KHX
 
 | |
1PXA
 
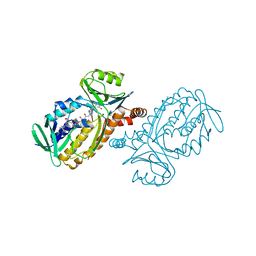 | | CRYSTAL STRUCTURES OF MUTANT PSEUDOMONAS AERUGINOSA P-HYDROXYBENZOATE HYDROXYLASE: THE TYR201PHE, TYR385PHE, AND ASN300ASP VARIANTS | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Lah, M.S, Palfey, B.A, Schreuder, H.A, Ludwig, M.L. | | Deposit date: | 1994-09-27 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of mutant Pseudomonas aeruginosa p-hydroxybenzoate hydroxylases: the Tyr201Phe, Tyr385Phe, and Asn300Asp variants.
Biochemistry, 33, 1994
|
|
1PXC
 
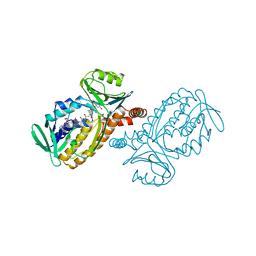 | | CRYSTAL STRUCTURES OF MUTANT PSEUDOMONAS AERUGINOSA P-HYDROXYBENZOATE HYDROXYLASE: THE TYR201PHE, TYR385PHE, AND ASN300ASP VARIANTS | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Lah, M.S, Palfey, B.A, Schreuder, H.A, Ludwig, M.L. | | Deposit date: | 1994-09-27 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of mutant Pseudomonas aeruginosa p-hydroxybenzoate hydroxylases: the Tyr201Phe, Tyr385Phe, and Asn300Asp variants.
Biochemistry, 33, 1994
|
|
3QY9
 
 | |
2BSL
 
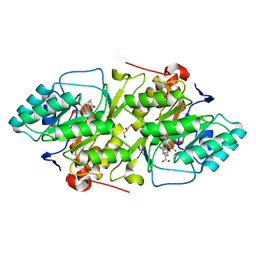 | | Crystal structure of L. lactis dihydroorotate dehydrogense A in complex with 3,4-dihydroxybenzoate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, ACETATE ION, DIHYDROOROTATE DEHYDROGENASE A, ... | | Authors: | Wolfe, A.E, Hansen, M, Gattis, S.G, Hu, Y.-C, Johansson, E, Arent, S, Larsen, S, Palfey, B.A. | | Deposit date: | 2005-05-23 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Interaction of Benzoate Pyrimidine Analogues with Class 1A Dihydroorotate Dehydrogenase from Lactococcus Lactis.
Biochemistry, 46, 2007
|
|
2BX7
 
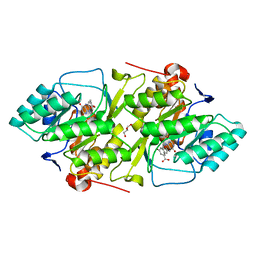 | | Crystal structure of L. lactis dihydroorotate dehydrogense A in complex with 3,5-dihydroxybenzoate | | Descriptor: | 3,5-DIHYDROXYBENZOATE, ACETATE ION, DIHYDROOROTATE DEHYDROGENASE, ... | | Authors: | Wolfe, A.E, Hansen, M, Gattis, S.G, Hu, Y.-C, Johansson, E, Arent, S, Larsen, S, Palfey, B.A. | | Deposit date: | 2005-07-25 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Interaction of Benzoate Pyrimidine Analogues with Class 1A Dihydroorotate Dehydrogenase from Lactococcus Lactis.
Biochemistry, 46, 2007
|
|
1PXB
 
 | | CRYSTAL STRUCTURES OF MUTANT PSEUDOMONAS AERUGINOSA P-HYDROXYBENZOATE HYDROXYLASE: THE TYR201PHE, TYR385PHE, AND ASN300ASP VARIANTS | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Lah, M.S, Palfey, B.A, Schreuder, H.A, Ludwig, M.L. | | Deposit date: | 1994-09-27 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of mutant Pseudomonas aeruginosa p-hydroxybenzoate hydroxylases: the Tyr201Phe, Tyr385Phe, and Asn300Asp variants.
Biochemistry, 33, 1994
|
|
1JF7
 
 | | HUMAN PTP1B CATALYTIC DOMAIN COMPLEXED WITH PNU177836 | | Descriptor: | 5-(2-{2-[(TERT-BUTOXY-HYDROXY-METHYL)-AMINO]-1-HYDROXY-3-PHENYL-PROPYLAMINO}-3-HYDROXY-3-PENTYLAMINO-PROPYL)-2-CARBOXYMETHOXY-BENZOIC ACID, PROTEIN-TYROSINE PHOSPHATASE 1B | | Authors: | Larsen, S.D, Barf, T, Liljebris, C, May, P.D, Ogg, D, O'Sullivan, T.J, Palazuk, B.J, Schostarez, H.J, Stevens, F.C, Bleasdale, J.E. | | Deposit date: | 2001-06-20 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and biological activity of a novel class of small molecular weight peptidomimetic competitive inhibitors of protein tyrosine phosphatase 1B.
J.Med.Chem., 45, 2002
|
|
3KX0
 
 | | Crystal Structure of the PAS domain of Rv1364c | | Descriptor: | ISOPROPYL ALCOHOL, Uncharacterized protein Rv1364c/MT1410 | | Authors: | Jaiswal, R.K, Gopal, B. | | Deposit date: | 2009-12-02 | | Release date: | 2010-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of a PAS sensor domain in the Mycobacterium tuberculosis transcription regulator Rv1364c
Biochem.Biophys.Res.Commun., 398, 2010
|
|
4NQW
 
 | |
3KI9
 
 | |
3S3K
 
 | |
3HUM
 
 | |
2PI7
 
 | |
3Q8Y
 
 | |
3HUN
 
 | |
3HUG
 
 | |
4EWT
 
 | | The crystal structure of a putative aminohydrolase from methicillin resistant Staphylococcus aureus | | Descriptor: | 1-DEOXY-1-THIO-HEPTAETHYLENE GLYCOL, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Girish, T.S, Vivek, B, Colaco, M, Misquith, S, Gopal, B. | | Deposit date: | 2012-04-27 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of an amidohydrolase, SACOL0085, from methicillin-resistant Staphylococcus aureus COL
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3Q86
 
 | |
5XE7
 
 | |
3Q83
 
 | |
3Q8V
 
 | |
