2F4W
 
 | | Human ubiquitin-conjugating enzyme E2 J2 | | Descriptor: | ubiquitin-conjugating enzyme E2, J2 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Finerty Jr, P.J, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-24 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
6VAJ
 
 | | Crystal Structure Analysis of human PIN1 | | Descriptor: | 2-chloro-N-(2,2-dimethylpropyl)-N-[(3R)-1,1-dioxo-1lambda~6~-thiolan-3-yl]acetamide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION, ... | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-12-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Sulfopin is a covalent inhibitor of Pin1 that blocks Myc-driven tumors in vivo.
Nat.Chem.Biol., 17, 2021
|
|
1ZKC
 
 | | Crystal Structure of the cyclophiln_RING domain of human peptidylprolyl isomerase (cyclophilin)-like 2 isoform b | | Descriptor: | BETA-MERCAPTOETHANOL, Peptidyl-prolyl cis-trans isomerase like 2 | | Authors: | Walker, J.R, Davis, T, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-05-02 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases.
PLoS Biol., 8, 2010
|
|
5VSB
 
 | | Structure of DUB complex | | Descriptor: | 7-chloro-3-{[4-hydroxy-1-(3-phenylpropanoyl)piperidin-4-yl]methyl}quinazolin-4(3H)-one, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2017-05-11 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7.
Cell Chem Biol, 24, 2017
|
|
5VS6
 
 | | Structure of DUB complex | | Descriptor: | ACETATE ION, GLYCEROL, N-[3-({4-hydroxy-1-[(3R)-3-phenylbutanoyl]piperidin-4-yl}methyl)-4-oxo-3,4-dihydroquinazolin-7-yl]-3-(4-methylpiperazin-1-yl)propanamide, ... | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2017-05-11 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7.
Cell Chem Biol, 24, 2017
|
|
2Z5D
 
 | | Human ubiquitin-conjugating enzyme E2 H | | Descriptor: | SODIUM ION, Ubiquitin-conjugating enzyme E2 H | | Authors: | Bochkarev, A, Cui, H, Walker, J.R, Newman, E.M, Mackenzie, F, Battaile, K.P, Sundstrom, M, Arrowsmith, C, Edwards, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-06 | | Release date: | 2007-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
MOL.CELL PROTEOMICS, 11, 2012
|
|
5TWX
 
 | | Crystal Structure of BRD9 bromodomain | | Descriptor: | Bromodomain-containing protein 9, N-[6-({2-[(3S)-2,6-dioxopiperidin-3-yl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}oxy)hexyl]-2-(4-{2-[N-(1,1-dioxo-1lambda~6~-thian-4-yl)carbamimidoyl]-5-methyl-4-oxo-4,5-dihydrothieno[3,2-c]pyridin-7-yl}-2-methoxyphenoxy)acetamide | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Degradation of the BAF Complex Factor BRD9 by Heterobifunctional Ligands.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
3EBB
 
 | | PLAP/P97 complex | | Descriptor: | MAGNESIUM ION, PHOSPHOLIPASE A2-ACTIVATING PROTEIN, TRANSITIONAL ENDOPLASMIC RETICULUM ATPASE (TER ATP | | Authors: | Walker, J.R, Qiu, L, Akutsu, M, Slessarev, Y, Amaya, M.F, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-27 | | Release date: | 2009-02-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the PLAA/Ufd3-p97/Cdc48 complex.
J.Biol.Chem., 285, 2010
|
|
5TWL
 
 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | 9-(3,5-dichloro-4-hydroxyphenyl)-1-{trans-4-[(dimethylamino)methyl]cyclohexyl}-3,4-dihydropyrimido[5,4-c]quinolin-2(1H)-one, Maternal embryonic leucine zipper kinase | | Authors: | Seo, H.-S, Huang, H.-T, Gray, N.S, Dhe-Paganon, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
5TWZ
 
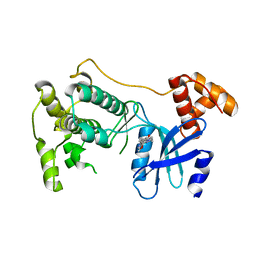 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | 7-{[2-methoxy-4-(1H-pyrazol-4-yl)benzoyl]amino}-2,3,4,5-tetrahydro-1H-3-benzazepinium, Maternal embryonic leucine zipper kinase | | Authors: | Seo, H.-Y, Dhe-Paganon, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
5TX3
 
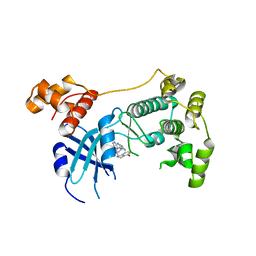 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | 7-[(1S)-4-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-2-({3-[(pyrrolidin-1-yl)methyl]phenyl}amino)-5,7-dihydro-6H-pyrrolo[2,3-d]pyrimidin-6-one, Maternal embryonic leucine zipper kinase | | Authors: | Li, Q, Seo, H.-S, Huang, H.-T, Gray, N.S, Dhe-Paganon, S, Eck, M.J. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
5TWY
 
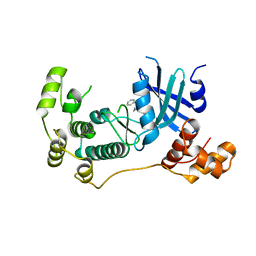 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | 2-(benzyloxy)-4-(1H-pyrazol-4-yl)-N-(2,3,4,5-tetrahydro-1H-3-benzazepin-7-yl)benzamide, Maternal embryonic leucine zipper kinase | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
5TWU
 
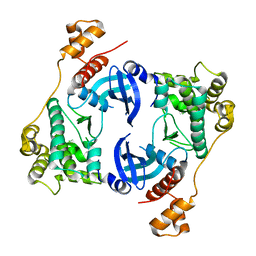 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | Maternal embryonic leucine zipper kinase | | Authors: | Li, Q, Seo, H.-S, Huang, H.-T, Gray, N.S, Dhe-Paganon, S, Eck, M.J. | | Deposit date: | 2016-11-14 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
3FL7
 
 | | Crystal structure of the human ephrin A2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Ephrin receptor, ... | | Authors: | Walker, J.R, Yermekbayeva, L, Seitova, A, Butler-Cole, C, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architecture of Eph receptor clusters.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3BI7
 
 | | Crystal structure of the SRA domain of E3 ubiquitin-protein ligase UHRF1 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, SULFATE ION, ... | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1.
Nature, 455, 2008
|
|
3BZH
 
 | | Crystal structure of human ubiquitin-conjugating enzyme E2 E1 | | Descriptor: | GLYCEROL, Ubiquitin-conjugating enzyme E2 E1 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-18 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
3ECI
 
 | | Microtubule-associated protein 1 light chain 3 alpha isoform A (MAP1ALC3) | | Descriptor: | Microtubule-associated protein 1 light chain 3 alpha | | Authors: | Walker, J.R, Davis, T.L, Mujib, S, Butler-Cole, C, Tempel, W, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Dhe-Paganon, S. | | Deposit date: | 2008-08-30 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Human Autophagy-Related Protein LC3 A
To be Published
|
|
3FXX
 
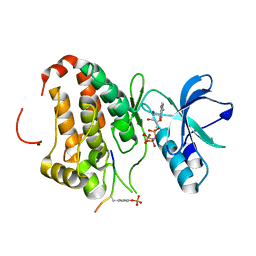 | | Human EphA3 Kinase and Juxtamembrane Region Bound to Substrate KQWDNYE[pTyr]IW | | Descriptor: | Ephrin type-A receptor 3, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Davis, T, Walker, J.R, Mackenzie, F, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-21 | | Release date: | 2009-02-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural recognition of an optimized substrate for the ephrin family of receptor tyrosine kinases.
Febs J., 276, 2009
|
|
3IHP
 
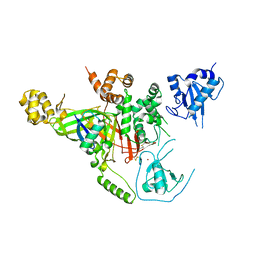 | | Covalent Ubiquitin-Usp5 Complex | | Descriptor: | CHLORIDE ION, ETHANAMINE, Ubiquitin, ... | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Butler-Cole, C, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-30 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Covalent Ubiquitin-Usp5 Complex
To be Published
|
|
3EPZ
 
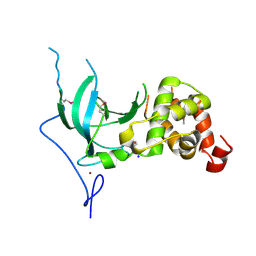 | | Structure of the replication foci-targeting sequence of human DNA cytosine methyltransferase DNMT1 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, GLYCEROL, SODIUM ION, ... | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The replication focus targeting sequence (RFTS) domain is a DNA-competitive inhibitor of Dnmt1.
J.Biol.Chem., 286, 2011
|
|
1ZIV
 
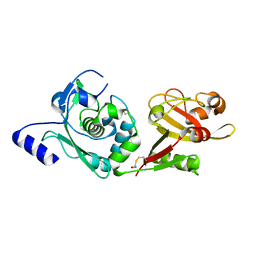 | | Catalytic Domain of Human Calpain-9 | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, Calpain 9 | | Authors: | Walker, J.R, Davis, T, Newman, E.M, Mackenzie, F, Dong, A, Choe, J, Arrowsmith, C, Sundstrom, M, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The crystal structures of human calpains 1 and 9 imply diverse mechanisms of action and auto-inhibition.
J.Mol.Biol., 366, 2007
|
|
3FL2
 
 | | Crystal structure of the ring domain of the E3 ubiquitin-protein ligase UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Ring Domain of the E3 Ubiquitin-Protein Ligase Uhrf1
To be Published
|
|
3DL2
 
 | | Hexagonal structure of the LDH domain of Human Ubiquitin-conjugating Enzyme E2-like Isoform A | | Descriptor: | PHOSPHATE ION, SODIUM ION, Ubiquitin-conjugating enzyme E2 variant 3 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Finerty Jr, P.J, Butler-Cole, C, Bountra, C, Wolkstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-26 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Investigation Into the L-Lactate Dehydrogenase Domain of Human Ubiquitin-Conjugating Enzyme E2-Like Isoform A.
To be Published
|
|
3H8H
 
 | | Structure of the C-terminal domain of human RNF2/RING1B; | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RING2, GLYCEROL, ... | | Authors: | Walker, J.R, Bezsonova, I, Bacik, J, Duan, S, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-29 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ring1B contains a ubiquitin-like docking module for interaction with Cbx proteins.
Biochemistry, 48, 2009
|
|
1ZKH
 
 | | Solution structure of a human ubiquitin-like domain in SF3A1 | | Descriptor: | Splicing factor 3 subunit 1 | | Authors: | Lukin, J.A, Dhe-Paganon, S, Guido, V, Lemak, A, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Sundstrom, M, Edwards, A, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-05-02 | | Release date: | 2005-05-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a human ubiquitin-like domain in SF3A1
To be Published
|
|
