3TPF
 
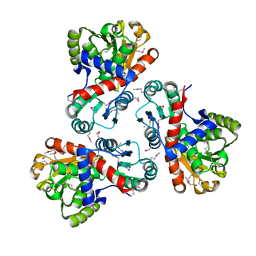 | | Crystal structure of anabolic ornithine carbamoyltransferase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ornithine carbamoyltransferase | | Authors: | Shabalin, I.G, Onopriyenko, O, Grimshaw, S, Porebski, P.J, Grabowski, M, Savchenko, A, Chruszcz, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-07 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of anabolic ornithine carbamoyltransferase from Campylobacter jejuni at 2.7 A resolution.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1KI2
 
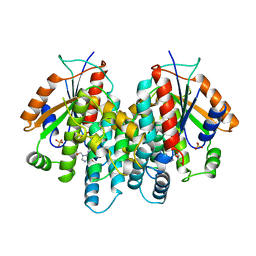 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH GANCICLOVIR | | Descriptor: | 9-(1,3-DIHYDROXY-PROPOXYMETHANE)GUANINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Brown, D.G, Visse, R, Sandhu, G, Davies, A, Rizkallah, P.J, Melitz, C, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-15 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
7Y7L
 
 | | Solution structure of zinc finger domain 2 of human ZFAND1 | | Descriptor: | AN1-type zinc finger protein 1, ZINC ION | | Authors: | Fang, P.J, Lai, C.H, Ko, K.T, Chang, C.F, Hsu, S.T.D. | | Deposit date: | 2022-06-22 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of p97 recognition by human ZFAND1
To Be Published
|
|
7YAB
 
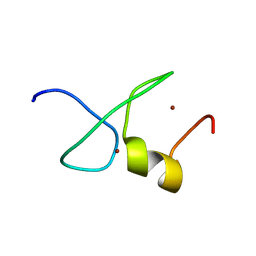 | | Solution structure of zinc finger domain 1 of human ZFAND1 | | Descriptor: | AN1-type zinc finger protein 1, ZINC ION | | Authors: | Fang, P.J, Lai, C.H, Ko, K.T, Chang, C.F, Hsu, S.T.D. | | Deposit date: | 2022-06-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of p97 recognition by human ZFAND1
To Be Published
|
|
3SX0
 
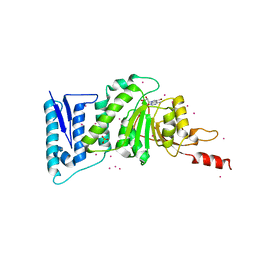 | | Crystal structure of Dot1l in complex with a brominated SAH analog | | Descriptor: | (2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(4-amino-5-bromo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}sulfanyl)butanoic acid (non-preferred name), Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yu, W, Tempel, W, Smil, D, Schapira, M, Li, Y, Vedadi, M, Nguyen, K.T, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-07-14 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Bromo-deaza-SAH: a potent and selective DOT1L inhibitor.
Bioorg. Med. Chem., 21, 2013
|
|
7Y39
 
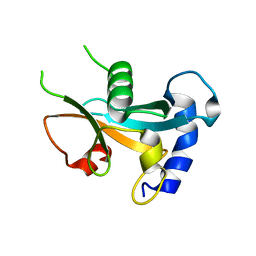 | | Ubiquitin-like domain of human ZFAND1 | | Descriptor: | AN1-type zinc finger protein 1 | | Authors: | Lai, C.H, Ko, K.T, Fan, P.J, Yu, T.A, Chang, C.F, Draczkowski, P, Hsu, S.T.D. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Insight into ZFAND1 and p97 Interaction
To Be Published
|
|
3QM1
 
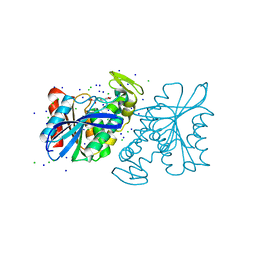 | | CRYSTAL STRUCTURE OF THE LACTOBACILLUS JOHNSONII CINNAMOYL ESTERASE LJ0536 S106A MUTANT IN COMPLEX WITH ETHYLFERULATE, Form II | | Descriptor: | CHLORIDE ION, Cinnamoyl esterase, SODIUM ION, ... | | Authors: | Stogios, P.J, Lai, K.K, Vu, C, Xu, X, Cui, H, Molloy, S, Gonzalez, C.F, Yakunin, A, Savchenko, A. | | Deposit date: | 2011-02-03 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.817 Å) | | Cite: | An Inserted alpha/beta Subdomain Shapes the Catalytic Pocket of Lactobacillus johnsonii Cinnamoyl Esterase
Plos One, 6, 2011
|
|
3Q6E
 
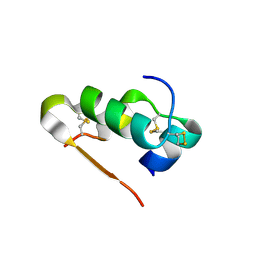 | | Human insulin in complex with cucurbit[7]uril | | Descriptor: | Insulin A chain, Insulin B chain, cucurbit[7]uril | | Authors: | Chinai, J.M, Taylor, A.B, Hargreaves, N.D, Ryno, L.M, Morris, C.A, Hart, P.J, Urbach, A.R. | | Deposit date: | 2010-12-31 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular recognition of insulin by a synthetic receptor.
J.Am.Chem.Soc., 133, 2011
|
|
3QQD
 
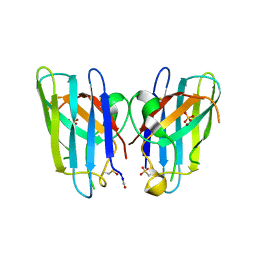 | | Human SOD1 H80R variant, P212121 crystal form | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Seetharaman, S.V, Winkler, D.D, Taylor, A.B, Cao, X, Whitson, L.J, Doucette, P.A, Valentine, J.S, Schirf, V, Demeler, B, Carroll, M.C, Culotta, V.C, Hart, P.J. | | Deposit date: | 2011-02-15 | | Release date: | 2011-03-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Disrupted zinc-binding sites in structures of pathogenic SOD1 variants D124V and H80R.
Biochemistry, 49, 2010
|
|
3TYK
 
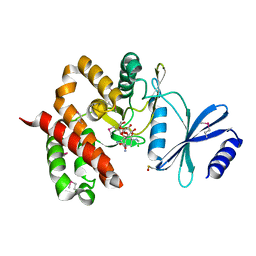 | | Crystal structure of aminoglycoside phosphotransferase APH(4)-Ia | | Descriptor: | CHLORIDE ION, HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase | | Authors: | Stogios, P.J, Shabalin, I.G, Shakya, T, Evdokmova, E, Fan, Y, Chruszcz, M, Minor, W, Wright, G.D, Savchenko, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and function of APH(4)-Ia, a hygromycin B resistance enzyme.
J.Biol.Chem., 286, 2011
|
|
3RPI
 
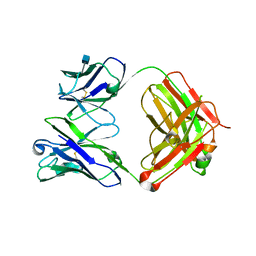 | | Crystal Structure of Fab from 3BNC60, Highly Potent anti-HIV Antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain from highly potent anti-HIV neutralizing antibody, Light chain from highly potent anti-HIV neutralizing antibody | | Authors: | Diskin, R, Bjorkman, P.J. | | Deposit date: | 2011-04-26 | | Release date: | 2011-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding.
Science, 333, 2011
|
|
3SMQ
 
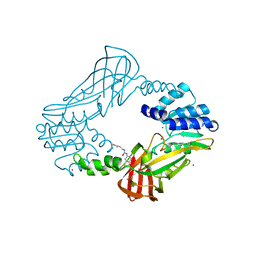 | | Crystal structure of protein arginine methyltransferase 3 | | Descriptor: | 1-(1,2,3-benzothiadiazol-6-yl)-3-[2-(cyclohex-1-en-1-yl)ethyl]urea, CHLORIDE ION, Protein arginine N-methyltransferase 3, ... | | Authors: | Dobrovetsky, E, Dong, A, Walker, J.R, Siarheyeva, A, Senisterra, G, Wasney, G.A, Smil, D, Bolshan, Y, Nguyen, K.T, Allali-Hassani, A, Hajian, T, Poda, G, Bountra, C, Weigelt, J, Edwards, A.M, Al-Awar, R, Brown, P.J, Schapira, M, Arrowsmith, C.H, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An allosteric inhibitor of protein arginine methyltransferase 3.
Structure, 20, 2012
|
|
3S2Z
 
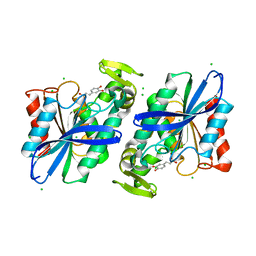 | | Crystal structure of the Lactobacillus johnsonii cinnamoyl esterase LJ0536 S106A mutant in complex with caffeic acid | | Descriptor: | CAFFEIC ACID, CHLORIDE ION, Cinnamoyl esterase | | Authors: | Stogios, P.J, Lai, K.K, Vu, C, Xu, X, Cui, H, Molloy, S, Gonzalez, C.F, Yakunin, A, Savchenko, A. | | Deposit date: | 2011-05-17 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An inserted alpha/beta subdomain shapes the catalytic pocket of Lactobacillus johnsonii cinnamoyl esterase.
Plos One, 6, 2011
|
|
3TLS
 
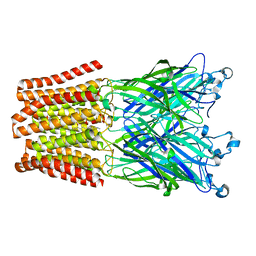 | | The GLIC pentameric Ligand-Gated Ion Channel E19'P mutant in a locally-closed conformation (LC2 subtype) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-08-30 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3TLU
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-24' oxidized mutant in a locally-closed conformation (LC1 subtype) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-08-30 | | Release date: | 2012-05-16 | | Last modified: | 2012-06-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2L4K
 
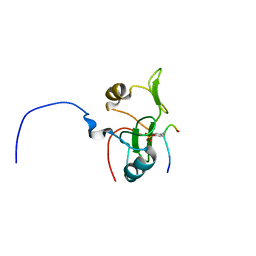 | | Water refined solution structure of the human Grb7-SH2 domain in complex with the 10 amino acid peptide pY1139 | | Descriptor: | Growth factor receptor-bound protein 7, Receptor tyrosine-protein kinase erbB-2 | | Authors: | Pias, S.C, Ivancic, M, Brescia, P.J, Johnson, D.L, Smith, D.E, Daly, R.J, Lyons, B.A. | | Deposit date: | 2010-10-07 | | Release date: | 2010-11-17 | | Last modified: | 2012-10-03 | | Method: | SOLUTION NMR | | Cite: | Water-Refined Solution Structure of the Human Grb7-SH2 Domain in Complex with the erbB2 Receptor Peptide pY1139.
Protein Pept.Lett., 19, 2012
|
|
3QE5
 
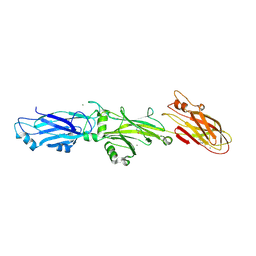 | | Complete structure of Streptococcus mutans Antigen I/II carboxy-terminus | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Major cell-surface adhesin PAc, ... | | Authors: | Larson, M.R, Rajashankar, K.R, Crowley, P.J, Kelly, C, Mitchell, T.J, Brady, L.J, Deivanayagam, C. | | Deposit date: | 2011-01-19 | | Release date: | 2011-04-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the C-terminal Region of Streptococcus mutans Antigen I/II and Characterization of Salivary Agglutinin Adherence Domains.
J.Biol.Chem., 286, 2011
|
|
3Q7E
 
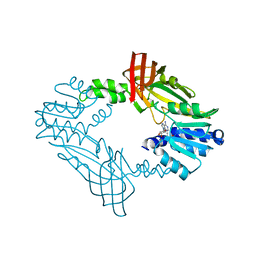 | |
3TLT
 
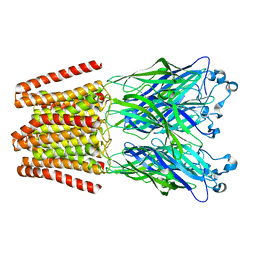 | | The GLIC pentameric Ligand-Gated Ion Channel H11'F mutant in a locally-closed conformation (LC1 subtype) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-08-30 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3TLV
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-22' oxidized mutant in a locally-closed conformation (LC3 subtype) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-08-30 | | Release date: | 2012-05-16 | | Last modified: | 2012-06-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3TYS
 
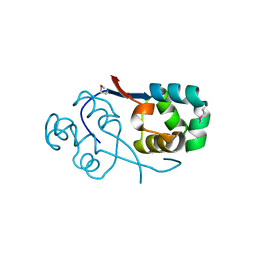 | | Crystal structure of transcriptional regulator VanUg, Form II | | Descriptor: | Predicted transcriptional regulator | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Depardieu, F, Courvalin, P, Shabalin, I, Chruszcz, M, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.121 Å) | | Cite: | Crystal structure of transcriptional regulator VanUg, Form II
TO BE PUBLISHED
|
|
3TYR
 
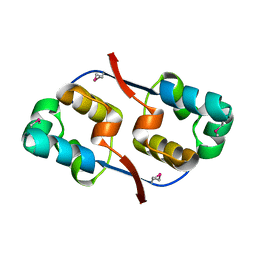 | | Crystal structure of transcriptional regulator VanUg, Form I | | Descriptor: | Transcriptional regulator | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Dong, A, Depardieu, F, Courvalin, P, Shabalin, I, Chruszcz, M, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of transcriptional regulator VanUg, Form I
TO BE PUBLISHED
|
|
3U7Y
 
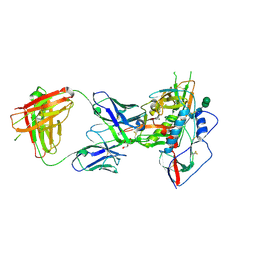 | |
3U7W
 
 | | Crystal structure of NIH45-46 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Heavy chain, ... | | Authors: | Diskin, R, Bjorkman, P.J. | | Deposit date: | 2011-10-14 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Increasing the Potency and Breadth of an HIV Antibody by Using Structure-Based Rational Design.
Science, 334, 2011
|
|
3UEG
 
 | | Crystal structure of human Survivin K62A mutant | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, TETRAETHYLENE GLYCOL, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
