1BDM
 
 | |
1BMD
 
 | |
7OL3
 
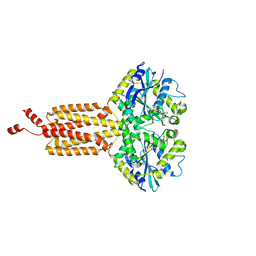 | | Human ATL1 N417ins (catalytic core) | | Descriptor: | Atlastin-1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kelly, C.M, Sondermann, H. | | Deposit date: | 2021-05-19 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel insertion mutation in atlastin 1 is associated with spastic quadriplegia, increased membrane tethering, and aberrant conformational switching.
J.Biol.Chem., 298, 2021
|
|
1Z7S
 
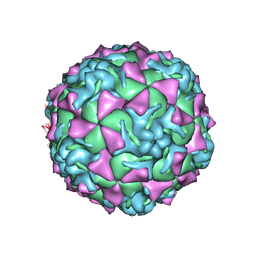 | | The crystal structure of coxsackievirus A21 | | Descriptor: | CALCIUM ION, GUANOSINE-5'-MONOPHOSPHATE, Human COXSACKIEVIRUS A21, ... | | Authors: | Xiao, C, Bator-Kelly, C.M, Rieder, E, Chipman, P.R, Craig, A, Kuhn, R.J, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2005-03-28 | | Release date: | 2005-08-02 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of coxsackievirus a21 and its interaction with icam-1.
Structure, 13, 2005
|
|
3QE5
 
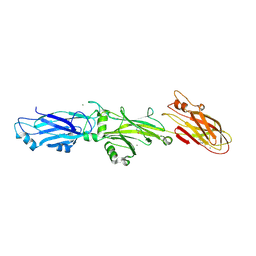 | | Complete structure of Streptococcus mutans Antigen I/II carboxy-terminus | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Major cell-surface adhesin PAc, ... | | Authors: | Larson, M.R, Rajashankar, K.R, Crowley, P.J, Kelly, C, Mitchell, T.J, Brady, L.J, Deivanayagam, C. | | Deposit date: | 2011-01-19 | | Release date: | 2011-04-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the C-terminal Region of Streptococcus mutans Antigen I/II and Characterization of Salivary Agglutinin Adherence Domains.
J.Biol.Chem., 286, 2011
|
|
1Z7Z
 
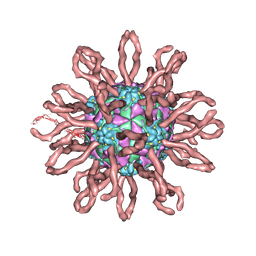 | | Cryo-em structure of human coxsackievirus A21 complexed with five domain icam-1kilifi | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Intercellular adhesion molecule-1, human coxsackievirus A21 | | Authors: | Xiao, C, Bator-Kelly, C.M, Rieder, E, Chipman, P.R, Craig, A, Kuhn, R.J, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2005-03-28 | | Release date: | 2005-08-02 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The crystal structure of coxsackievirus a21 and its interaction with icam-1.
Structure, 13, 2005
|
|
2B6B
 
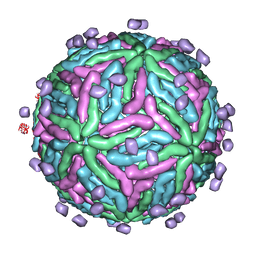 | | Cryo EM structure of Dengue complexed with CRD of DC-SIGN | | Descriptor: | CD209 antigen, envelope glycoprotein | | Authors: | Pokidysheva, E, Zhang, Y, Battisti, A.J, Bator-Kelly, C.M, Chipman, P.R, Gregorio, G, Hendrickson, W.A, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2005-09-30 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN
Cell(Cambridge,Mass.), 124, 2006
|
|
2QZH
 
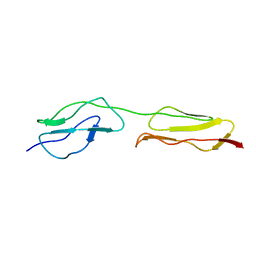 | | SCR2/3 of DAF from the NMR structure 1nwv fitted into a cryoEM reconstruction of CVB3-RD complexed with DAF | | Descriptor: | Complement decay-accelerating factor | | Authors: | Hafenstein, S, Bowman, V.D, Chipman, P.R, Bator Kelly, C.M, Lin, F, Medof, M.E, Rossmann, M.G. | | Deposit date: | 2007-08-16 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Interaction of decay-accelerating factor with coxsackievirus b3.
J.Virol., 81, 2007
|
|
2QZF
 
 | | SCR1 of DAF from 1ojv fitted into cryoEM density | | Descriptor: | Complement decay-accelerating factor | | Authors: | Hafenstein, S, Bowman, V.D, Chipman, P.R, Bator Kelly, C.M, Lin, F, Medof, M.E, Rossmann, M.G. | | Deposit date: | 2007-08-16 | | Release date: | 2007-10-30 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Interaction of decay-accelerating factor with coxsackievirus b3.
J.Virol., 81, 2007
|
|
2QZD
 
 | | Fitted structure of SCR4 of DAF into cryoEM density | | Descriptor: | Complement decay-accelerating factor | | Authors: | Hafenstein, S, Bowman, V.D, Chipman, P.R, Bator Kelly, C.M, Lin, F, Medof, M.E, Rossmann, M.G. | | Deposit date: | 2007-08-16 | | Release date: | 2007-10-30 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Interaction of decay-accelerating factor with coxsackievirus b3.
J.Virol., 81, 2007
|
|
