8OTV
 
 | | Crystal structure of NUDT14 complexed with novel compound | | Descriptor: | 1-(1-methylpiperidin-4-yl)-3-(4-phenoxyphenyl)pyrazolo[3,4-d]pyrimidin-4-amine, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Balikci, E, Feyerherm, C, Bradshaw, W, Seupel, R, Brennan, P.E, Bountra, C, von Delft, F, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Unexpected Noncovalent Off-Target Activity of Clinical BTK Inhibitors Leads to Discovery of a Dual NUDT5/14 Antagonist.
J.Med.Chem., 67, 2024
|
|
6T1N
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 5 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
7MON
 
 | | Structure of human RIPK3-MLKL complex | | Descriptor: | Mixed lineage kinase domain-like protein, N-[4-({2-[(cyclopropanecarbonyl)amino]pyridin-4-yl}oxy)-3-fluorophenyl]-1-(4-fluorophenyl)-2-oxo-1,2-dihydropyridine-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Meng, Y, Davies, K.A, Czabotar, P.E, Murphy, J.M. | | Deposit date: | 2021-05-03 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Human RIPK3 maintains MLKL in an inactive conformation prior to cell death by necroptosis.
Nat Commun, 12, 2021
|
|
6T1O
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 6 | | Descriptor: | 1,2-ETHANEDIOL, 4-iodanyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
7MX3
 
 | | Crystal structure of human RIPK3 complexed with GSK'843 | | Descriptor: | 1,2-ETHANEDIOL, 3-(1,3-benzothiazol-5-yl)-7-(1,3-dimethyl-1H-pyrazol-5-yl)thieno[3,2-c]pyridin-4-amine, Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Davies, K.A, Czabotar, P.E. | | Deposit date: | 2021-05-18 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Human RIPK3 maintains MLKL in an inactive conformation prior to cell death by necroptosis.
Nat Commun, 12, 2021
|
|
3D7V
 
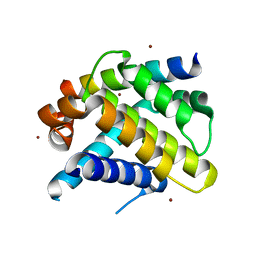 | | Crystal structure of Mcl-1 in complex with an Mcl-1 selective BH3 ligand | | Descriptor: | Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Lee, E.F, Czabotar, P.E, Colman, P.M, Fairlie, W.D. | | Deposit date: | 2008-05-22 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A novel BH3 ligand that selectively targets Mcl-1 reveals that apoptosis can proceed without Mcl-1 degradation.
J.Cell Biol., 180, 2008
|
|
4HTP
 
 | |
4HTO
 
 | |
4ICD
 
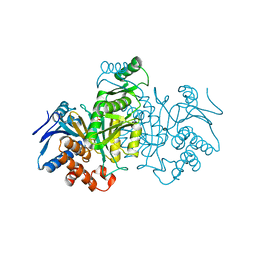 | | REGULATION OF ISOCITRATE DEHYDROGENASE BY PHOSPHORYLATION INVOLVES NO LONG-RANGE CONFORMATIONAL CHANGE IN THE FREE ENZYME | | Descriptor: | PHOSPHORYLATED ISOCITRATE DEHYDROGENASE | | Authors: | Hurley, J.H, Dean, A.M, Thorsness, P.E, Koshlandjunior, D.E, Stroud, R.M. | | Deposit date: | 1989-12-28 | | Release date: | 1991-01-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of isocitrate dehydrogenase by phosphorylation involves no long-range conformational change in the free enzyme.
J.Biol.Chem., 265, 1990
|
|
4NX7
 
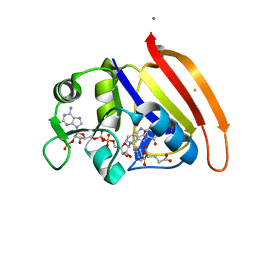 | | single cryogenic temperature model of DHFR | | Descriptor: | BETA-MERCAPTOETHANOL, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | Fenwick, R.B, van den Bedem, H, Fraser, J.S, Wright, P.E. | | Deposit date: | 2013-12-08 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Integrated description of protein dynamics from room-temperature X-ray crystallography and NMR.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NX6
 
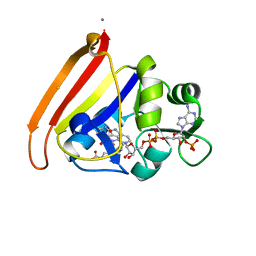 | | single room temperature model of DHFR | | Descriptor: | BETA-MERCAPTOETHANOL, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | Fenwick, R.B, van den Bedem, H, Fraser, J.S, Wright, P.E. | | Deposit date: | 2013-12-08 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Integrated description of protein dynamics from room-temperature X-ray crystallography and NMR.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NXJ
 
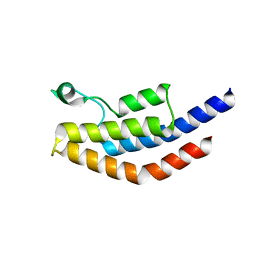 | | Crystal Structure of PF3D7_1475600, a bromodomain from Plasmodium Falciparum | | Descriptor: | Bromodomain protein | | Authors: | Wernimont, A.K, Loppnau, P, Knapp, S, Fonseca, M, Brennan, P.E, Dong, A, Walker, J.R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-09 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of PF3D7_1475600, a bromodomain from Plasmodium Falciparum
TO BE PUBLISHED
|
|
4M6K
 
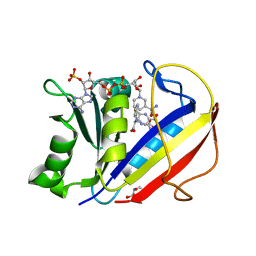 | | Crystal structure of human dihydrofolate reductase (DHFR) bound to NADP+ and folate | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, GLYCEROL, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Divergent evolution of protein conformational dynamics in dihydrofolate reductase.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4KJJ
 
 | | Cryogenic WT DHFR | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | van den Bedem, H, Bhabha, G, Yang, K, Wright, P.E, Fraser, J.S. | | Deposit date: | 2013-05-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Automated identification of functional dynamic contact networks from X-ray crystallography.
Nat.Methods, 10, 2013
|
|
4KJL
 
 | | Room Temperature N23PPS148A DHFR | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | van den Bedem, H, Bhabha, G, Yang, K, Wright, P.E, Fraser, J.S. | | Deposit date: | 2013-05-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Automated identification of functional dynamic contact networks from X-ray crystallography.
Nat.Methods, 10, 2013
|
|
4MWI
 
 | | Crystal structure of the human MLKL pseudokinase domain | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, GLYCEROL, Mixed lineage kinase domain-like protein | | Authors: | Czabotar, P.E, Murphy, J.M. | | Deposit date: | 2013-09-25 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into the evolution of divergent nucleotide-binding mechanisms among pseudokinases revealed by crystal structures of human and mouse MLKL.
Biochem.J., 457, 2014
|
|
4PY6
 
 | | Crystal Structure of bromodomain of PFA0510w from Plasmodium Falciparum | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Bromodomain protein, ... | | Authors: | Fonseca, M, Tallant, C, Hutchinson, A, Savitsky, P, Krojer, T, Filippakopoulos, P, Loppnau, P, Brennan, P.E, von Delft, F, Dong, A, Josling, G.A, Duffy, M.F, Arrowsmith, C.H, Bountra, C, Hui, R, Knapp, S, Wernimont, A.K, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of bromodomain of PFA0510w from Plasmodium Falciparum
To be Published
|
|
4KJK
 
 | | Room Temperature WT DHFR | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | van den Bedem, H, Bhabha, G, Yang, K, Wright, P.E, Fraser, J.S. | | Deposit date: | 2013-05-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Automated identification of functional dynamic contact networks from X-ray crystallography.
Nat.Methods, 10, 2013
|
|
9PCY
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF REDUCED FRENCH BEAN PLASTOCYANIN AND COMPARISON WITH THE CRYSTAL STRUCTURE OF POPLAR PLASTOCYANIN | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Moore, J.M, Lepre, C.A, Gippert, G.P, Chazin, W.J, Case, D.A, Wright, P.E. | | Deposit date: | 1991-03-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of reduced French bean plastocyanin and comparison with the crystal structure of poplar plastocyanin.
J.Mol.Biol., 221, 1991
|
|
3UBI
 
 | | The Absence of Tertiary Interactions in a Self-Assembled DNA Crystal Structure | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*CP*GP*TP*AP*CP*TP*CP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*AP*GP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*CP*GP*AP*GP*TP*AP*CP*GP*AP*CP*GP*AP*CP*AP*AP*G)-3'), ... | | Authors: | Nguyen, N, Birktoft, J.J, Sha, R, Wang, T, Zheng, J, Constantinou, P.E, Ginell, S.L, Chen, Y, Mao, C, Seeman, N.C. | | Deposit date: | 2011-10-24 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (6.8046 Å) | | Cite: | The absence of tertiary interactions in a self-assembled DNA crystal structure.
J.Mol.Recognit., 25, 2012
|
|
5CCC
 
 | |
6MCY
 
 | | Crystal structure of mouse Bak | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2 homologous antagonist/killer, FORMIC ACID | | Authors: | Brouwer, J.M, Czabotar, P.E, Colman, P.M. | | Deposit date: | 2018-09-03 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | A small molecule interacts with VDAC2 to block mouse BAK-driven apoptosis.
Nat.Chem.Biol., 15, 2019
|
|
2YQ7
 
 | | Structure of Bcl-xL bound to BimLOCK | | Descriptor: | BCL-2-LIKE PROTEIN 1, BCL-2-LIKE PROTEIN 11, GLYCEROL | | Authors: | Smith, B.J, Czabotar, P.E. | | Deposit date: | 2012-11-06 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Stabilizing the Pro-Apoptotic Bimbh3 Helix (Bimsahb) Does not Necessarily Enhance Affinity or Biological Activity.
Acs Chem.Biol., 8, 2013
|
|
3S2Y
 
 | | Crystal structure of a chromate/uranium reductase from Gluconacetobacter hansenii | | Descriptor: | CHLORIDE ION, Chromate reductase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Jin, H, Zhang, Y, Buchko, G.W, Li, P, Squier, T.C, Robinson, H, Varnum, S.M, Long, P.E. | | Deposit date: | 2011-05-17 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Structure Determination and Functional Analysis of a Chromate Reductase from Gluconacetobacter hansenii.
Plos One, 7, 2012
|
|
2VOH
 
 | | Structure of mouse A1 bound to the Bak BH3-domain | | Descriptor: | BCL-2 HOMOLOGOUS ANTAGONIST/KILLER, BCL-2-RELATED PROTEIN A1, CITRIC ACID, ... | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
