2QUE
 
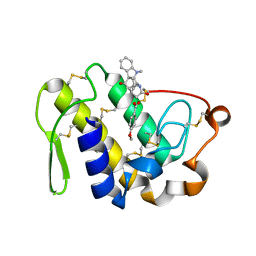 | | Saturation of substrate-binding site using two natural ligands: Crystal structure of a ternary complex of phospholipase A2 with anisic acid and ajmaline at 2.25 A resolution | | Descriptor: | 4-METHOXYBENZOIC ACID, AJMALINE, Phospholipase A2 VRV-PL-VIIIa | | Authors: | Kumar, S, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-08-05 | | Release date: | 2007-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Saturation of substrate-binding site using two natural ligands: Crystal structure of a ternary complex of phospholipase A2 with anisic acid and ajmaline at 2.25 A resolution
To be Published
|
|
1M1A
 
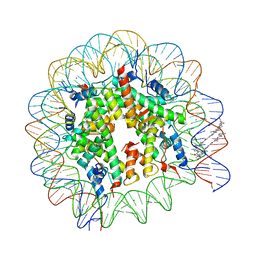 | | LIGAND BINDING ALTERS THE STRUCTURE AND DYNAMICS OF NUCLEOSOMAL DNA | | Descriptor: | 3-AMINO-(DIMETHYLPROPYLAMINE), 4-AMINO-(1-METHYLIMIDAZOLE)-2-CARBOXYLIC ACID, 4-AMINO-(1-METHYLPYRROLE)-2-CARBOXYLIC ACID, ... | | Authors: | Suto, R.K, Edayathumangalam, R.S, White, C.L, Melander, C, Gottesfeld, J.M, Dervan, P.B, Luger, K. | | Deposit date: | 2002-06-18 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of Nucleosome Core Particles in Complex with Minor Groove DNA-binding Ligands
J.MOL.BIOL., 326, 2003
|
|
1M3X
 
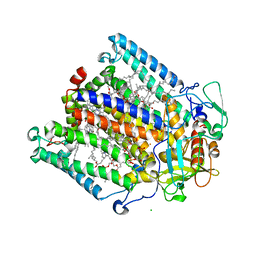 | | Photosynthetic Reaction Center From Rhodobacter Sphaeroides | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Camara-Artigas, A, Brune, D, Allen, J.P. | | Deposit date: | 2002-07-01 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Interactions between lipids and bacterial reaction centers determined by protein crystallography.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3QGK
 
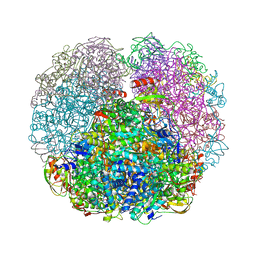 | |
1QPY
 
 | | CRYSTAL STRUCTURE OF BACKBONE MODIFIED PNA HEXAMER | | Descriptor: | PEPTIDE NUCLEIC ACID 5'-(*CP1*GPN*TP1*APN*CP1*GPN*LYS)-3' | | Authors: | Haima, G, Rasmussen, H, Schmidt, G, Jensen, D.K, Kastrup, J.S, Stafshede, P.W, Norden, B, Buchardt, O, Nielsen, P.E. | | Deposit date: | 1999-05-14 | | Release date: | 2001-02-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peptide Nucleic Acids (PNA) derived from N-(N-methylaminoethyl)glycine. Synthesis, hybridization and structural properties
New J.Chem., 23, 1999
|
|
1ZP5
 
 | | Crystal structure of the complex between MMP-8 and a N-hydroxyurea inhibitor | | Descriptor: | CALCIUM ION, N-{2-[(4'-CYANO-1,1'-BIPHENYL-4-YL)OXY]ETHYL}-N'-HYDROXY-N-METHYLUREA, Neutrophil collagenase, ... | | Authors: | Campestre, C, Agamennone, M, Tortorella, P, Preziuso, S, Biasone, A, Gavuzzo, E, Pochetti, G, Mazza, F, Tschesche, H, Gallina, C. | | Deposit date: | 2005-05-16 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | N-Hydroxyurea as zinc binding group in matrix metalloproteinase inhibition: Mode of binding in a complex with MMP-8.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4JQI
 
 | | Structure of active beta-arrestin1 bound to a G protein-coupled receptor phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-arrestin-1, CHLORIDE ION, ... | | Authors: | Shukla, A.K, Manglik, A, Kruse, A.C, Xiao, K, Reis, R.I, Tseng, W.C, Staus, D.P, Hilger, D, Uysal, S, Huang, L.H, Paduch, M, Shukla, P.T, Koide, A, Koide, S, Weis, W.I, Kossiakoff, A.A, Kobilka, B.K, Lefkowitz, R.J. | | Deposit date: | 2013-03-20 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of active beta-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide.
Nature, 497, 2013
|
|
4JV9
 
 | | Co-crystal structure of MDM2 with inhibitor (2S,5R,6S)-2-benzyl-5,6-bis(4-chlorophenyl)-4-methylmorpholin-3-one | | Descriptor: | (2S,5R,6S)-2-benzyl-5,6-bis(4-chlorophenyl)-4-methylmorpholin-3-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
1EHZ
 
 | |
2JES
 
 | | Portal protein (gp6) from bacteriophage SPP1 | | Descriptor: | CALCIUM ION, MERCURY (II) ION, PORTAL PROTEIN, ... | | Authors: | Lebedev, A.A, Krause, M.H, Isidro, A.L, Vagin, A.A, Orlova, E.V, Turner, J, Dodson, E.J, Tavares, P, Antson, A.A. | | Deposit date: | 2007-01-21 | | Release date: | 2007-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Framework for DNA Translocation Via the Viral Portal Protein
Embo J., 26, 2007
|
|
1QXR
 
 | | Crystal structure of phosphoglucose isomerase from Pyrococcus furiosus in complex with 5-phosphoarabinonate | | Descriptor: | 5-PHOSPHOARABINONIC ACID, Glucose-6-phosphate isomerase, NICKEL (II) ION | | Authors: | Swan, M.K, Solomons, J.T.G, Beeson, C.C, Hansen, P, Schonheit, P, Davies, C. | | Deposit date: | 2003-09-08 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural evidence for a hydride transfer mechanism of catalysis in phosphoglucose isomerase from Pyrococcus furiosus
J.Biol.Chem., 278, 2003
|
|
1QY7
 
 | | The structure of the PII protein from the cyanobacteria Synechococcus sp. PCC 7942 | | Descriptor: | NICKEL (II) ION, Nitrogen regulatory protein P-II, SULFATE ION | | Authors: | Xu, Y, Carr, P.D, Clancy, P, Garcia-Dominguez, M, Forchhammer, K, Florencio, F, Tandeau de Marsac, N, Vasudevan, S.G, Ollis, D.L. | | Deposit date: | 2003-09-09 | | Release date: | 2003-09-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of the PII proteins from the cyanobacteria Synechococcus sp. PCC 7942 and Synechocystis sp. PCC 6803.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4AYW
 
 | | STRUCTURE OF THE HUMAN MITOCHONDRIAL ABC TRANSPORTER, ABCB10 (PLATE FORM) | | Descriptor: | ATP-BINDING CASSETTE SUB-FAMILY B MEMBER 10, CHOLESTEROL HEMISUCCINATE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pike, A.C.W, Shintre, C.A, Li, Q, Kim, J, von Delft, F, Barr, A.J, Das, S, Chaikuad, A, Xia, X, Quigley, A, Dong, Y, Dong, L, Krojer, T, Vollmar, M, Muniz, J.R.C, Bray, J.E, Berridge, G, Chalk, R, Gileadi, O, Burgess-Brown, N, Shrestha, L, Goubin, S, Yang, J, Mahajan, P, Mukhopadhyay, S, Bullock, A.N, Arrowsmith, C.H, Weigelt, J, Bountra, C, Edwards, A.M, Carpenter, E.P. | | Deposit date: | 2012-06-22 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Abcb10, a Human ATP-Binding Cassette Transporter in Apo- and Nucleotide-Bound States
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2FZ5
 
 | | Solution structure of two-electron reduced Megasphaera elsdenii flavodoxin | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Flavodoxin | | Authors: | van Mierlo, C.P.M, Lijnzaad, P, Vervoort, J, Mueller, F, Berendsen, H.J, de Vlieg, J. | | Deposit date: | 2006-02-09 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Tertiary structure of two-electron reduced Megasphaera elsdenii flavodoxin and some implications, as determined by two-dimensional 1H NMR and restrained molecular dynamics
Eur.J.Biochem., 194, 1990
|
|
4JDO
 
 | | Secreted chlamydial protein pgp3, coiled-coil deletion | | Descriptor: | SODIUM ION, Virulence plasmid protein pGP3-D | | Authors: | Galaleldeen, A, Taylor, A.B, Chen, D, Holloway, S.P, Zhong, G, Hart, P.J. | | Deposit date: | 2013-02-25 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Chlamydia trachomatis immunodominant antigen Pgp3.
J.Biol.Chem., 288, 2013
|
|
2A2Q
 
 | | Complex of Active-site Inhibited Human Coagulation Factor VIIa with Human Soluble Tissue Factor in the Presence of Ca2+, Mg2+, Na+, and Zn2+ | | Descriptor: | CALCIUM ION, CHLORIDE ION, Coagulation factor VII, ... | | Authors: | Bajaj, S.P, Bajaj, M, Schmidt, A.E, Padmanabhan, K. | | Deposit date: | 2005-06-22 | | Release date: | 2006-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structures of p-aminobenzamidine- and benzamidine-VIIa/soluble tissue factor: unpredicted conformation of the 192-193 peptide bond and mapping of Ca2+, Mg2+, Na+, and Zn2+ sites in factor VIIa.
J.Biol.Chem., 281, 2006
|
|
4BIE
 
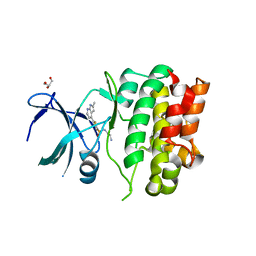 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, N-(2-aminoethyl)-5-{2-methyl-1H-pyrrolo[2,3-b]pyridin-4-yl}thiophene-2-sulfonamide | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
3U7Y
 
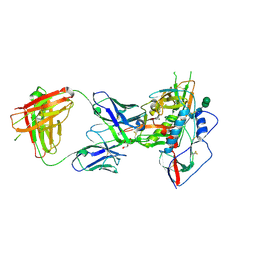 | |
3U7W
 
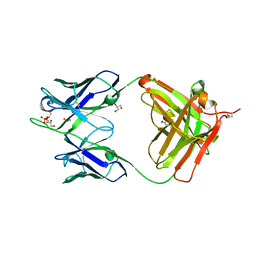 | | Crystal structure of NIH45-46 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Heavy chain, ... | | Authors: | Diskin, R, Bjorkman, P.J. | | Deposit date: | 2011-10-14 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Increasing the Potency and Breadth of an HIV Antibody by Using Structure-Based Rational Design.
Science, 334, 2011
|
|
3TWM
 
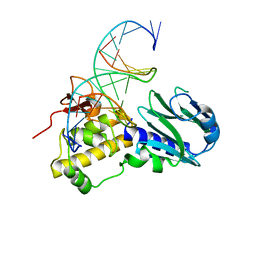 | | Crystal structure of Arabidopsis thaliana FPG | | Descriptor: | 5'-D(*AP*GP*CP*GP*TP*CP*CP*AP*(3DR)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*TP*GP*GP*TP*AP*GP*AP*CP*GP*TP*GP*GP*AP*CP*GP*C)-3', Formamidopyrimidine-DNA glycosylase 1 | | Authors: | Duclos, S, Aller, P, Wallace, S.S, Doublie, S. | | Deposit date: | 2011-09-22 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical studies of a plant formamidopyrimidine-DNA glycosylase reveal why eukaryotic Fpg glycosylases do not excise 8-oxoguanine.
Dna Repair, 11, 2012
|
|
4K4Y
 
 | | Coxsackievirus B3 polymerase elongation complex (r2+1_form) | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, ACETATE ION, DNA/RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*(DOC))-3'), ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2013-04-12 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts.
Plos One, 8, 2013
|
|
2FVZ
 
 | | Human Inositol Monophosphosphatase 2 | | Descriptor: | Inositol monophosphatase 2 | | Authors: | Ogg, D, Hallberg, B.M, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Graslund, S, Hammarstrom, M, Hogbom, M, Holmberg-Schiavone, L, Kotenyova, T, Kursula, P, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Van Den Berg, S, Weigelt, J, Thorsell, A.G, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-31 | | Release date: | 2006-02-21 | | Last modified: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Human Inositol Monophosphatase 2
To be published
|
|
2LLJ
 
 | | Structure of a bis-naphthalene bound to a thymine-thymine DNA mismatch | | Descriptor: | 6,22-dioxa-3,9,19,25-tetraazoniapentacyclo[25.5.3.3~11,17~.0~14,37~.0~30,34~]octatriaconta-1(33),11(38),12,14(37),15,17(36),27,29,31,34-decaene, DNA (5'-D(*CP*GP*TP*CP*GP*TP*AP*GP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*CP*TP*TP*CP*GP*AP*CP*G)-3') | | Authors: | Jourdan, M, Granzhan, A, Guillot, R, Dumy, P, Teulade-Fichou, M. | | Deposit date: | 2011-11-10 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Double threading through DNA: NMR structural study of a bis-naphthalene macrocycle bound to a thymine-thymine mismatch.
Nucleic Acids Res., 40, 2012
|
|
2LAC
 
 | | NMR structure of unmodified_ASL_Tyr | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*AP*CP*UP*GP*UP*AP*AP*AP*UP*CP*CP*CP*C)-3') | | Authors: | Denmon, A.P, Wang, J, Nikonowicz, E.P. | | Deposit date: | 2011-03-10 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformation Effects of Base Modification on the Anticodon Stem-Loop of Bacillus subtilis tRNA(Tyr).
J.Mol.Biol., 412, 2011
|
|
2M5Q
 
 | |
