4DOE
 
 | | The liganded structure of Cbescii CelA GH9 module | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 1,4-beta-glucanase, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2012-02-09 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Revealing nature's cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA.
Science, 342, 2013
|
|
7RS3
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-29 | | Descriptor: | (1S,2R,4S)-6-[4-(benzyloxy)phenyl]-5-(4-hydroxyphenyl)-N-(4-methoxyphenyl)-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, CYSTEINE, Estrogen receptor, ... | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5O8A
 
 | | Crystal Structure of rsEGFP2 in the non-fluorescent off-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
4DT4
 
 | |
2HL0
 
 | | Crystal structure of the editing domain of threonyl-tRNA synthetase from Pyrococcus abyssi in complex with seryl-3'-aminoadenosine | | Descriptor: | SERINE-3'-AMINOADENOSINE, Threonyl-tRNA synthetase | | Authors: | Hussain, T, Kruparani, S.P, Pal, B, Sankaranarayanan, R. | | Deposit date: | 2006-07-06 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Post-transfer editing mechanism of a D-aminoacyl-tRNA deacylase-like domain in threonyl-tRNA synthetase from archaea
Embo J., 25, 2006
|
|
5O4Y
 
 | | Structure of human PD-L1 in complex with inhibitor | | Descriptor: | PHE-MAA-ASN-PRO-HIS-LEU-SER-TRP-SER-TRP-9KK-9KK-ARG-CCS-GLY-NH2, Programmed cell death 1 ligand 1 | | Authors: | Magiera, K, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-05-31 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bioactive Macrocyclic Inhibitors of the PD-1/PD-L1 Immune Checkpoint.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5O9O
 
 | | Crystal structure of ScGas2 in complex with compound 7. | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-[4-(naphthalen-2-ylmethoxymethyl)-1,2,3-triazol-1-yl]-3,5-bis(oxidanyl)oxan-4-yl]oxy-3,5-bis(oxidanyl)oxan-4-yl]oxy-oxane-3,4,5-triol, 1,2-ETHANEDIOL, 1,3-beta-glucanosyltransferase GAS2 | | Authors: | Delso, I, Valero-Gonzalez, J, Fang, W, Gomollon-Bel, F, Castro-Lopez, J, Navratilova, I, van Aalten, D, Tejero, T, Merino, P, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-19 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibitors against Fungal Cell Wall Remodeling Enzymes.
ChemMedChem, 13, 2018
|
|
5O9Y
 
 | | Crystal structure of ScGas2 in complex with compound 11 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-3,5-bis(oxidanyl)-6-[4-(3-pyridin-1-ium-1-ylpropyl)-1,2,3-triazol-1-yl]oxan-4-yl]oxy-3,5-bis(oxidanyl)oxan-4-yl]oxy-oxane-3,4,5-triol, 1,2-ETHANEDIOL, 1,3-beta-glucanosyltransferase GAS2, ... | | Authors: | Delso, I, Valero-Gonzalez, J, Gomollon-Bel, F, Castro-Lopez, J, Fang, W, Navratilova, I, Van Aalten, D, Tejero, T, Merino, P, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Inhibitors against Fungal Cell Wall Remodeling Enzymes.
ChemMedChem, 13, 2018
|
|
4DE4
 
 | | Crystal structure of aminoglycoside phosphotransferase APH(2")-Id/APH(2")-IVa in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, APH(2")-Id | | Authors: | Stogios, P.J, Minasov, G, Tan, K, Nocek, B, Evdokimova, E, Egorova, O, Di Leo, R, Li, H, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-19 | | Release date: | 2012-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A small molecule discrimination map of the antibiotic resistance kinome.
Chem.Biol., 18, 2011
|
|
8AK4
 
 | | Structure of the C-terminally truncated NAD+-dependent DNA ligase from the poly-extremophile Deinococcus radiodurans | | Descriptor: | DNA ligase, MANGANESE (II) ION, ZINC ION | | Authors: | Fernandes, A, Williamson, A.K, Matias, P.M, Moe, E. | | Deposit date: | 2022-07-29 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structure/function studies of the NAD + -dependent DNA ligase from the poly-extremophile Deinococcus radiodurans reveal importance of the BRCT domain for DNA binding.
Extremophiles, 27, 2023
|
|
2KYZ
 
 | | NMR structure of heavy metal binding protein TM0320 from Thermotoga maritima | | Descriptor: | Heavy metal binding protein | | Authors: | Jaudzems, K, Wahab, A, Serrano, P, Geralt, M, Wuthrich, K, Wilson, I.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-06-09 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of heavy metal binding protein TM0320 from Thermotoga maritima
To be Published
|
|
3G80
 
 | |
3G8C
 
 | | Crystal Structure of Biotin Carboxylase in Complex with Biotin, Bicarbonate, ADP and Mg Ion | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BICARBONATE ION, BIOTIN, ... | | Authors: | Chou, C.Y, Yu, L.P, Tong, L. | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of biotin carboxylase in complex with substrates and implications for its catalytic mechanism.
J.Biol.Chem., 284, 2009
|
|
4DGI
 
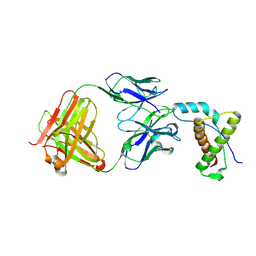 | | Structure of POM1 FAB fragment complexed with human PrPc Fragment 120-230 | | Descriptor: | Major prion protein, POM1 Fab Heavy chain, POM1 Fab Light chain, ... | | Authors: | Baral, P.K, Wieland, B, Swayampakula, M, James, M.N. | | Deposit date: | 2012-01-26 | | Release date: | 2012-10-31 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies on the folded domain of the human prion protein bound to the Fab fragment of the antibody POM1.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2KUK
 
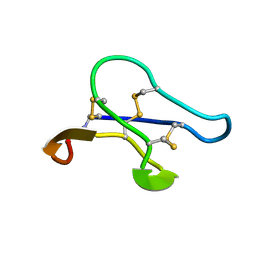 | |
1VRY
 
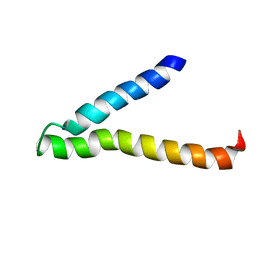 | | Second and Third Transmembrane Domains of the Alpha-1 Subunit of Human Glycine Receptor | | Descriptor: | Glycine receptor alpha-1 chain | | Authors: | Ma, D, Liu, Z, Li, L, Tang, P, Xu, Y. | | Deposit date: | 2005-07-20 | | Release date: | 2005-07-26 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Second and Third Transmembrane Domains of Human Glycine Receptor.
Biochemistry, 44, 2005
|
|
3G9B
 
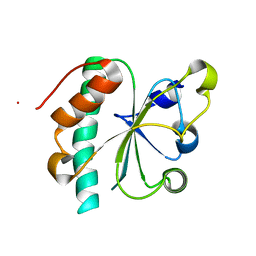 | | Crystal structure of reduced Ost6L | | Descriptor: | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6 | | Authors: | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | Deposit date: | 2009-02-13 | | Release date: | 2009-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GAI
 
 | | Structure of a F112A variant PduO-type ATP:corrinoid adenosyltransferase from Lactobacillus reuteri complexed with cobalamin and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, COBALAMIN, ... | | Authors: | St Maurice, M, Mera, P.E, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2009-02-17 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Residue Phe112 of the human-type corrinoid adenosyltransferase (PduO) enzyme of Lactobacillus reuteri is critical to the formation of the four-coordinate Co(II) corrinoid substrate and to the activity of the enzyme.
Biochemistry, 48, 2009
|
|
2KXC
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for IRTKS-SH3 and EspFu-R47 complex | | Descriptor: | Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1, EspF-like protein | | Authors: | Aitio, O, Hellman, M, Kazlauskas, A, Vingadassalom, D.F, Leong, J.M, Saksela, K, Permi, P. | | Deposit date: | 2010-04-30 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of tandem PxxP motifs as a unique Src homology 3-binding mode triggers pathogen-driven actin assembly
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4DYS
 
 | | Crystal Structure of Apo Swine Flu Influenza Nucleoprotein | | Descriptor: | Nucleocapsid protein | | Authors: | Lewis, H.A, Baldwin, E.T, Steinbacher, S, Maskos, K, Mortl, M, Kiefersauer, R, Edavettal, S, McDonnell, P.A, Pearce, B.C, Langley, D.R. | | Deposit date: | 2012-02-29 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | To be determined
To be Published
|
|
4E0H
 
 | | Crystal structure of FAD binding domain of Erv1 from Saccharomyces cerevisiae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Mitochondrial FAD-linked sulfhydryl oxidase ERV1 | | Authors: | Guo, P.C, Ma, J.D, Jiang, Y.L, Wang, S.J, Hu, T.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2012-03-04 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of yeast sulfhydryl oxidase erv1 reveals electron transfer of the disulfide relay system in the mitochondrial intermembrane space
J.Biol.Chem., 287, 2012
|
|
7RBS
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with human ISG15 | | Descriptor: | Papain-like protease, Ubiquitin-like protein ISG15, ZINC ION | | Authors: | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Wydorski, P, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
4DN8
 
 | | Structure of porcine surfactant protein D neck and carbohydrate recognition domain complexed with mannose | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein D, beta-D-mannopyranose | | Authors: | van Eijk, M, Rynkiewicz, M.J, White, M.R, Hartshorn, K.L, Zou, X, Schulten, K, Luo, D, Crouch, E.C, Cafarella, T.M, Head, J.F, Haagsman, H.P, Seaton, B.A. | | Deposit date: | 2012-02-08 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Unique Sugar-binding Site Mediates the Distinct Anti-influenza Activity of Pig Surfactant Protein D.
J.Biol.Chem., 287, 2012
|
|
4DOI
 
 | |
4DPZ
 
 | | Crystal structure of human HRASLS2 | | Descriptor: | HRAS-like suppressor 2 | | Authors: | Kiser, P.D, Golczak, M, Sears, A.E, Lodowski, D.T, Palczewski, K. | | Deposit date: | 2012-02-14 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis for the Acyltransferase Activity of Lecithin:Retinol Acyltransferase-like Proteins.
J.Biol.Chem., 287, 2012
|
|
