4DIG
 
 | | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with N-acetylmuramyl l-alanyl d-isoglutamine at 1.8 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shukla, P.K, Gautam, L, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-01-31 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with N-acetylmuramyl l-alanyl d-isoglutamine at 1.8 A Resolution
To be Published
|
|
3GCK
 
 | | Mode of ligand binding and assignment of subsites in mammalian peroxidases: crystal structure of lactoperoxidase complexes with acetyl salycylic acid, salicylhydroxamic acid and benzylhydroxamic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BENZHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Singh, A.K, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mode of ligand binding and assignment of subsites in mammalian peroxidases: crystal structure of lactoperoxidase complexes with acetyl salycylic acid, salicylhydroxamic acid and benzylhydroxamic acid
To be Published
|
|
3MNG
 
 | | wild type human PrxV with DTT bound as a competitive inhibitor | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, BROMIDE ION, GLYCEROL, ... | | Authors: | Hall, A, Karplus, P.A. | | Deposit date: | 2010-04-21 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Evidence that Peroxiredoxin Catalytic Power Is Based on Transition-State Stabilization.
J.Mol.Biol., 402, 2010
|
|
6O25
 
 | | Crystal structure of 3945 Fab in complex with circumsporozoite protein NANP3 and anti-Kappa VHH domain | | Descriptor: | 3945 Fab heavy chain, 3945 Kappa light chain, Anti-Kappa VHH domain, ... | | Authors: | Scally, S.W, Bosch, A, Prieto, K, Murugan, R, Wardemann, H, Julien, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs.
Nat. Med., 26, 2020
|
|
6XNR
 
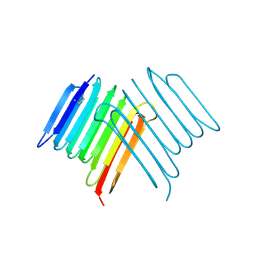 | | Crystal structure of Rhagium Mordax antifreeze protein | | Descriptor: | 1,2-ETHANEDIOL, Antifreeze protein | | Authors: | Ye, Q, Eves, R, Campbell, R.L, Davies, P.L. | | Deposit date: | 2020-07-04 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of an insect antifreeze protein reveals ordered waters on the ice-binding surface.
Biochem.J., 477, 2020
|
|
6NKE
 
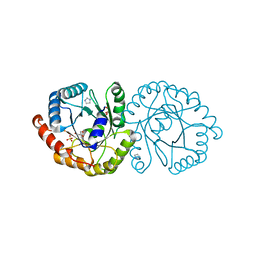 | | Wild-type GGGPS from Thermoplasma volcanium | | Descriptor: | GLYCEROL, Geranylgeranylglyceryl phosphate synthase, IMIDAZOLE, ... | | Authors: | Blank, P.N, Alderfer, K.E, Gillott, B.N, Christianson, D.W, Himmelberger, J.A. | | Deposit date: | 2019-01-07 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural studies of geranylgeranylglyceryl phosphate synthase, a prenyltransferase found in thermophilic Euryarchaeota.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6L2J
 
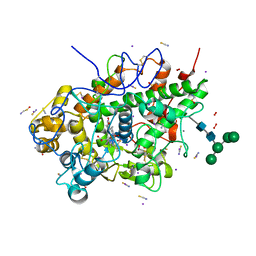 | | Crystal structure of yak lactoperoxidase at 1.93 A resolution. | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Viswanathan, V, Sharma, P, Rani, C, Sharma, S, Singh, T.P. | | Deposit date: | 2019-10-04 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Crystal structure of yak lactoperoxidase at 1.93 A resolution.
To Be Published
|
|
6XK0
 
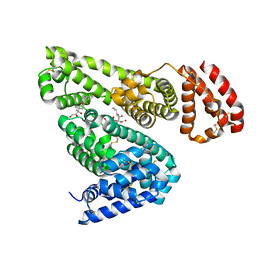 | | Albumin-dexamethasone complex | | Descriptor: | Albumin, CITRATE ANION, DEXAMETHASONE, ... | | Authors: | Czub, M.P, Majorek, K.A, Shabalin, I.G, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2020-06-24 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular determinants of vascular transport of dexamethasone in COVID-19 therapy.
Iucrj, 7, 2020
|
|
6NLQ
 
 | |
6XKF
 
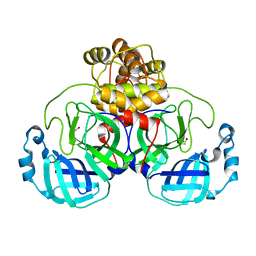 | | The crystal structure of 3CL MainPro of SARS-CoV-2 with oxidized Cys145 (Sulfenic acid cysteine). | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Coates, L, Kovalevskyi, A.Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of 3CL MainPro of SARS-CoV-2 with oxidized Cys145 (Sulfenic acid cysteine).
To Be Published
|
|
1RM5
 
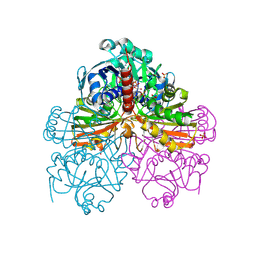 | | Crystal structure of mutant S188A of photosynthetic glyceraldehyde-3-phosphate dehydrogenase A4 isoform, complexed with NADP | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase A, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Sparla, F, Fermani, S, Falini, G, Ripamonti, A, Sabatino, P, Pupillo, P, Trost, P. | | Deposit date: | 2003-11-27 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Coenzyme Site-directed Mutants of Photosynthetic A(4)-GAPDH Show Selectively Reduced NADPH-dependent Catalysis, Similar to Regulatory AB-GAPDH Inhibited by Oxidized Thioredoxin
J.Mol.Biol., 340, 2004
|
|
4OW3
 
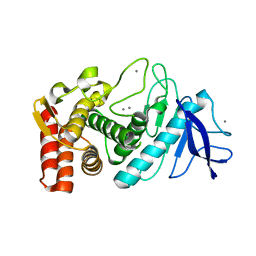 | | Thermolysin structure determined by free-electron laser | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Hattne, J, Echols, N, Tran, R, Kern, J, Gildea, R.J, Brewster, A.S, Alonso-Mori, R, Glockner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, White, W.E, Schafer, D.W, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Glatzel, P, Zwart, P.H, Grosse-Kunstleve, R.W, Bogan, M.J, Messerschmidt, M, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Yano, J, Bergmann, U, Yachandra, V.K, Adams, P.D, Sauter, N.K. | | Deposit date: | 2014-01-30 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Accurate macromolecular structures using minimal measurements from X-ray free-electron lasers.
Nat.Methods, 11, 2014
|
|
6L9N
 
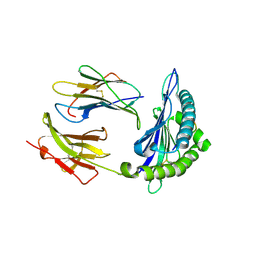 | | H2-Ld complexed with A5 peptide | | Descriptor: | MHC, SER-PRO-SER-TYR-ALA-TYR-HIS-GLN-PHE, b2m | | Authors: | Wei, P.C, Yin, L. | | Deposit date: | 2019-11-10 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures suggest an approach for converting weak self-peptide tumor antigens into superagonists for CD8 T cells in cancer.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5O7Y
 
 | | Thebaine 6-O-demethylase (T6ODM) from Papaver somniferum in complex with succinate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICKEL (II) ION, ... | | Authors: | Kluza, A, Niedzialkowska, E, Kurpiewska, K, Porebski, P.J, Borowski, T. | | Deposit date: | 2017-06-10 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of thebaine 6-O-demethylase from the morphine biosynthesis pathway.
J. Struct. Biol., 202, 2018
|
|
3M58
 
 | | SET7/9 Y245A in complex with TAF10-K189me1 peptide and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
5OA0
 
 | | Crystal structure of mutant AChBP in complex with strychnine (T53F, Q74R, Y110A, I135S, W164F) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, STRYCHNINE, ... | | Authors: | Dawson, A, Hunter, W.N, de Souza, J.O, Trumper, P. | | Deposit date: | 2017-06-20 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering a surrogate human heteromeric alpha / beta glycine receptor orthosteric site exploiting the structural homology and stability of acetylcholine-binding protein.
Iucrj, 6, 2019
|
|
6LCT
 
 | | Crystal structure of catalytic inactive chloroplast resolvase NtMOC1 in complex with Holliday junction | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*TP*GP*TP*CP*CP*CP*TP*CP*TP*GP*TP*TP*GP*T)-3'), DNA (5'-D(*AP*CP*AP*AP*CP*AP*GP*AP*GP*GP*AP*TP*GP*GP*AP*GP*CP*T)-3'), DNA (5'-D(*GP*CP*CP*TP*TP*GP*CP*TP*GP*GP*GP*AP*CP*AP*TP*CP*TP*T)-3'), ... | | Authors: | Yan, J.J, Hong, S.X, Guan, Z.Y, Yin, P. | | Deposit date: | 2019-11-19 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1.
Nat Commun, 11, 2020
|
|
5OAL
 
 | | Crystal structure of mutant AChBP in complex with strychnine (T53F, Q74R, Y110A, I135S, G162E) | | Descriptor: | ARGININE, STRYCHNINE, Soluble acetylcholine receptor | | Authors: | Dawson, A, Hunter, W.N, de Souza, J.O, Trumper, P. | | Deposit date: | 2017-06-22 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Engineering a surrogate human heteromeric alpha / beta glycine receptor orthosteric site exploiting the structural homology and stability of acetylcholine-binding protein.
Iucrj, 6, 2019
|
|
5OAN
 
 | | Crystal structure of mutant AChBP in complex with glycine (T53F, Q74R, Y110A, I135S, G162E, S206CCP_KGTG) | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCINE, ... | | Authors: | Dawson, A, Hunter, W.N, de Souza, J.O, Trumper, P. | | Deposit date: | 2017-06-23 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering a surrogate human heteromeric alpha / beta glycine receptor orthosteric site exploiting the structural homology and stability of acetylcholine-binding protein.
Iucrj, 6, 2019
|
|
6NR6
 
 | | Crystal Structure of Staphylococcus pseudintermedius SbnI in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional transcriptional regulator/O-phospho-L-serine synthase SbnI, MAGNESIUM ION | | Authors: | Verstraete, M.M, Morales, L.D, Kobylarz, M.J, Loutet, S.A, Laakso, H.A, Heinrichs, D.E, Murphy, M.E.P. | | Deposit date: | 2019-01-22 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The heme-sensitive regulator SbnI has a bifunctional role in staphyloferrin B production by Staphylococcus aureus .
J.Biol.Chem., 294, 2019
|
|
3M61
 
 | | Crystal structure of complex of urokinase and a upain-1 variant(W3A) in pH4.6 condition | | Descriptor: | Urokinase-type plasminogen activator, upain-1 W3A | | Authors: | Jiang, L, Yuan, C, Wind, T, Andreasen, P.A, Chen, L, Meehan, E.J, Huang, M. | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of complex of urokinase and a upain-1 variant(W3A) in pH4.6 condition
TO BE PUBLISHED
|
|
3M52
 
 | | Crystal structure of the BTB domain from the Miz-1/ZBTB17 transcription regulator | | Descriptor: | ACETATE ION, ZINC ION, Zinc finger and BTB domain-containing protein 17 | | Authors: | Stogios, P.J, Cuesta-Seijo, J.A, Chen, L, Prive, G.G. | | Deposit date: | 2010-03-12 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Insights into Strand Exchange in BTB Domain Dimers from the Crystal Structures of FAZF and Miz1.
J.Mol.Biol., 400, 2010
|
|
3M5A
 
 | | SET7/9 Y245A in complex with TAF10-K189me3 peptide and AdoHcy | | Descriptor: | COBALT (II) ION, GLYCEROL, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
2BOP
 
 | | CRYSTAL STRUCTURE AT 1.7 ANGSTROMS OF THE BOVINE PAPILLOMAVIRUS-1 E2 DNA-BINDING DOMAIN BOUND TO ITS DNA TARGET | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*CP*GP*AP*CP*GP*TP*CP*GP*GP*TP*CP*G )-3'), PROTEIN (E2), YTTERBIUM (III) ION | | Authors: | Hegde, R.S, Grossman, S.R, Laimins, L.A, Sigler, P.B. | | Deposit date: | 1994-01-13 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure at 1.7 A of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target.
Nature, 359, 1992
|
|
4YQI
 
 | | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds | | Descriptor: | (4-amino-1,2,5-oxadiazol-3-yl)[4-(pyridin-2-ylmethyl)piperazin-1-yl]methanone, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Elkins, P.A, Bonnette, W.G, Madauss, K.P, Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds
To Be Published
|
|
