5G2N
 
 | | X-ray structure of PI3Kinase Gamma in complex with Copanlisib | | Descriptor: | 2-azanyl-~{N}-[7-methoxy-8-(3-morpholin-4-ylpropoxy)-2,3-dihydroimidazo[1,2-c]quinazolin-5-yl]pyrimidine-5-carboxamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM, SULFATE ION | | Authors: | Schaefer, M, Scott, W.J, Hentemann, M.F, Rowley, R.B, Bull, C.O, Jenkins, S, Bullion, A.M, Johnson, J, Redman, A, Robbins, A.H, Esler, W, Fracasso, R.P, Garrison, T, Hamilton, M, Michels, M, Wood, J.E, Wilkie, D.P, Xiao, H, Levy, J, Liu, N, Stasik, E, Brands, M, Lefranc, J. | | Deposit date: | 2016-04-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery and Sar of Novel 2,3-Dihydroimidazo(1,2-C)Quinazoline Pi3K Inhibitors: Identification of Copanlisib (Bay 80-6946)
Chemmedchem, 11, 2016
|
|
6CPY
 
 | | Structure of apo GRMZM2G135359 pseudokinase | | Descriptor: | GRMZM2G135359 pseudokinase | | Authors: | Aquino, B, Counago, R.M, Godoi, P.H.C, Massirer, K.B, Elkins, J.M, Arruda, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-14 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of apo GRMZM2G135359 pseudokinase
To be Published
|
|
1JLC
 
 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-2 | | Descriptor: | HIV-1 RT A-chain, HIV-1 RT B-chain, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-CHLORO-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
6E52
 
 | |
5G1V
 
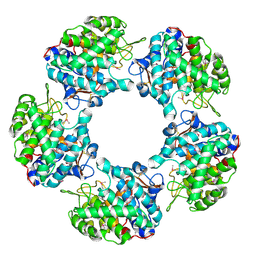 | | Linalool Dehydratase Isomerase: Selenomethionine Derivative | | Descriptor: | LINALOOL DEHYDRATASE ISOMERASE | | Authors: | Chambers, S, Hau, A, Man, H, Omar, M, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-03-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural and functional insights into asymmetric enzymatic dehydration of alkenols.
Nat. Chem. Biol., 13, 2017
|
|
4P23
 
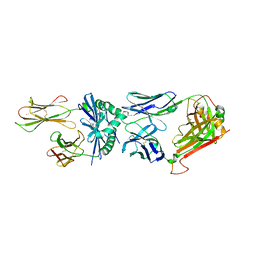 | | J809.B5 TCR bound to IAb/3K | | Descriptor: | 3K peptide and MHC IAb beta chain,H-2 class II histocompatibility antigen, A beta chain, H-2 class II histocompatibility antigen, ... | | Authors: | Stadinski, B.D, Huseby, E.S, Trenh, P, Stern, L. | | Deposit date: | 2014-02-28 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Effect of CDR3 Sequences and Distal V Gene Residues in Regulating TCR-MHC Contacts and Ligand Specificity.
J Immunol., 192, 2014
|
|
7N2A
 
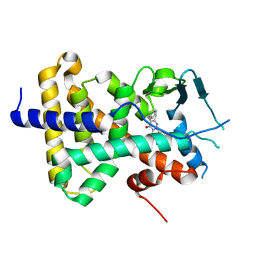 | | human PXR LBD bound to compound 2 | | Descriptor: | 5-benzyl-2-(3-fluoro-2-hydroxyphenyl)-6-methyl-3-(2-phenylethyl)pyrimidin-4(3H)-one, Isoform 1C of Nuclear receptor subfamily 1 group I member 2 | | Authors: | Williams, S.P, Wisely, G.B, Ramanjulu, J.M. | | Deposit date: | 2021-05-28 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Overcoming the Pregnane X Receptor Liability: Rational Design to Eliminate PXR-Mediated CYP Induction.
Acs Med.Chem.Lett., 12, 2021
|
|
5NBC
 
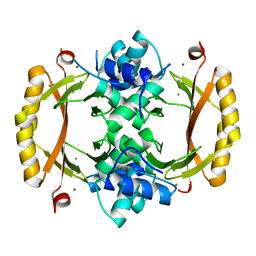 | | Structure of Prokaryotic Transcription Factors | | Descriptor: | Ferric uptake regulation protein, MANGANESE (II) ION, ZINC ION | | Authors: | Perard, J, Carpentier, P, Michaud-Soret, I, Cavazza, C. | | Deposit date: | 2017-03-01 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural and functional studies of the metalloregulator Fur identify a promoter-binding mechanism and its role inFrancisella tularensisvirulence.
Commun Biol, 1, 2018
|
|
1G0B
 
 | | CARBONMONOXY LIGANDED EQUINE HEMOGLOBIN PH 8.5 | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN ALPHA CHAIN, HEMOGLOBIN BETA CHAIN, ... | | Authors: | Mueser, T.C, Rogers, P.H, Arnone, A. | | Deposit date: | 2000-10-05 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interface sliding as illustrated by the multiple quaternary structures of liganded hemoglobin.
Biochemistry, 39, 2000
|
|
6ER0
 
 | | 6th KOW domain of human hSpt5 | | Descriptor: | Transcription elongation factor SPT5 | | Authors: | Hahn, L, Schweimer, K, Roesch, P, Woehrl, B.M. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and nucleic acid binding properties of KOW domains 4 and 6-7 of human transcription elongation factor DSIF.
Sci Rep, 8, 2018
|
|
6JAN
 
 | | Crystal structure of ABC transporter alpha-glycoside-binding mutant protein R356A in complex with maltose | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, periplasmic substrate-binding protein, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
4MH2
 
 | | Crystal structure of Bovine Mitochondrial Peroxiredoxin III | | Descriptor: | CITRIC ACID, Thioredoxin-dependent peroxide reductase, mitochondrial | | Authors: | Cao, Z, McGow, D.P, Shepherd, C, Lindsay, J.G. | | Deposit date: | 2013-08-29 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improved Catenated Structures of Bovine Peroxiredoxin III F190L Reveal Details of Ring-Ring Interactions and a Novel Conformational State.
Plos One, 10, 2015
|
|
5NDB
 
 | | Crystal structure of metallo-beta-lactamase SPM-1 complexed with cyclobutanone inhibitor | | Descriptor: | (1~{S},4~{R},5~{S})-7,7-bis(chloranyl)-6,6-bis(oxidanyl)-2$l^{4}-thiabicyclo[3.2.0]hept-2-ene-4-carboxylic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase IMP-1, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2017-03-08 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Cyclobutanone Mimics of Intermediates in Metallo-beta-Lactamase Catalysis.
Chemistry, 24, 2018
|
|
6J4G
 
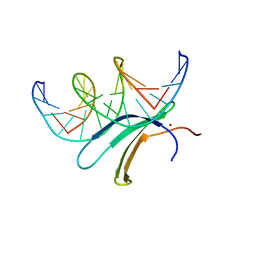 | | Crystal structure of the AtWRKY33 domain | | Descriptor: | DNA (5'-D(*AP*GP*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*C)-3'), Probable WRKY transcription factor 33, ... | | Authors: | Xu, Y.P, Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the N-terminal DNA binding domain of AtWRKY33
To Be Published
|
|
4PNS
 
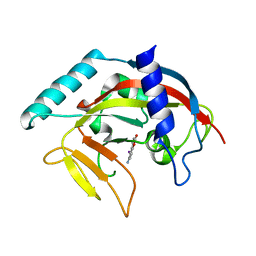 | | Crystal Structure of human Tankyrase 2 in complex with INH2BP. | | Descriptor: | 6-amino-5-iodo-2H-chromen-2-one, GLYCEROL, Tankyrase-2, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5KSR
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa - XFSurE-TB (Tetramer Bigger). | | Descriptor: | 5'-nucleotidase SurE, CHLORIDE ION, IODIDE ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, De Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
6IXN
 
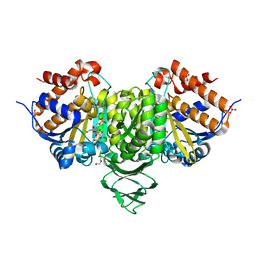 | | Crystal structure of isocitrate dehydrogenase from Ostreococcus tauri in complex with NAD+ and citrate | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Zhu, G.P, Tang, W.G, Wang, P. | | Deposit date: | 2018-12-11 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of NAD + -linked isocitrate dehydrogenase from the green alga Ostreococcus tauri and its evolutionary relationship with eukaryotic NADP + -linked homologs.
Arch.Biochem.Biophys., 708, 2021
|
|
1JQK
 
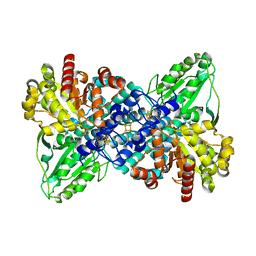 | | Crystal structure of carbon monoxide dehydrogenase from Rhodospirillum rubrum | | Descriptor: | FE (II) ION, FE(3)-NI(1)-S(4) CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Drennan, C.L, Heo, J, Sintchak, M.D, Schreiter, E, Ludden, P.W. | | Deposit date: | 2001-08-07 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Life on carbon monoxide: X-ray structure of Rhodospirillum rubrum Ni-Fe-S carbon monoxide dehydrogenase.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3LCX
 
 | | L-KDO aldolase | | Descriptor: | N-acetylneuraminate lyase, SULFATE ION | | Authors: | Chou, C.-Y, Huang, K.-F, Wang, A.H.-J, Ko, T.-P. | | Deposit date: | 2010-01-12 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Modulation of substrate specificities of D-sialic acid aldolase through single mutations of Val251
To be Published
|
|
5MFJ
 
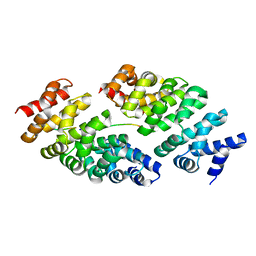 | | Designed armadillo repeat protein YIII(Dq.V2)4CqI in complex with peptide (KR)5 | | Descriptor: | (KR)5, YIII(Dq.V2)4CqI | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
8RBT
 
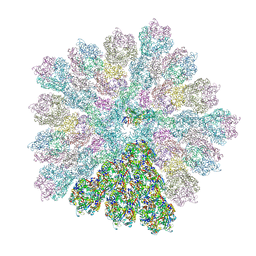 | | Emiliania huxleyi virus 201 (EhV-201) capsid proteins predicted by AlphaFold2 fitted into a cryo-EM density map of the EhV-201 virion capsid. | | Descriptor: | Major capsid protein, Penton protein | | Authors: | Homola, M, Buttner, C.R, Fuzik, T, Novacek, J, Chaillet, M, Forster, F, Plevka, P. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure and replication cycle of a virus infecting climate-modulating alga Emiliania huxleyi.
Sci Adv, 10, 2024
|
|
6VMS
 
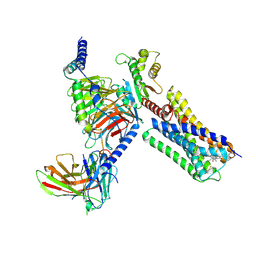 | | Structure of a D2 dopamine receptor-G-protein complex in a lipid membrane | | Descriptor: | Endolysin,D(2) dopamine receptor,D(2) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yin, J, Chen, K.M, Clark, M.J, Hijazi, M, Kumari, P, Bai, X, Sunahara, R.K, Barth, P, Rosenbaum, D.M. | | Deposit date: | 2020-01-28 | | Release date: | 2020-06-17 | | Last modified: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a D2 dopamine receptor-G-protein complex in a lipid membrane.
Nature, 584, 2020
|
|
6CU8
 
 | | Alpha Synuclein fibril formed by full length protein - Twister Polymorph | | Descriptor: | Alpha-synuclein | | Authors: | Li, B, Hatami, A, Ge, P, Murray, K.A, Sheth, P, Zhang, M, Nair, G, Sawaya, M.R, Zhu, C, Broad, M, Shin, W.S, Ye, S, John, V, Eisenberg, D.S, Zhou, Z.H, Jiang, L. | | Deposit date: | 2018-03-23 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM of full-length alpha-synuclein reveals fibril polymorphs with a common structural kernel.
Nat Commun, 9, 2018
|
|
7KBZ
 
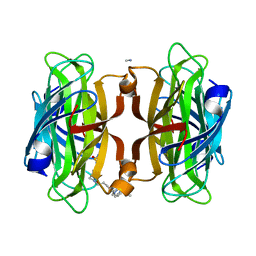 | | Artificial Metalloproteins with Dinuclear Iron Centers | | Descriptor: | CYANIDE ION, Streptavidin, {N-(4-{bis[(pyridin-2-yl-kappaN)methyl]amino-kappaN}butyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}iron(3+) | | Authors: | Miller, K.R, Follmer, A.H, Jasniewski, A.J, Sabuncu, S, Biswas, S, Albert, T, Hendrich, M.P, Moenne-Loccoz, P, Borovik, A.S. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Artificial Metalloproteins with Dinuclear Iron-Hydroxido Centers.
J.Am.Chem.Soc., 143, 2021
|
|
4TSH
 
 | | A Novel Protein Fold Forms an Intramolecular Lock to Stabilize the Tertiary Structure of Streptococcus mutans Adhesin P1 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Surface protein adhesin | | Authors: | Heim, K.P, Kailasan, S, McKenna, R, Brady, L.J. | | Deposit date: | 2014-06-18 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An intramolecular lock facilitates folding and stabilizes the tertiary structure of Streptococcus mutans adhesin P1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
